Figures & data
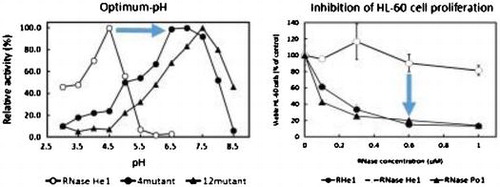
Fig. 1. Comparison of the amino acid sequences of RNase He1, RNase Po1, RNase T1, and RNase Ms.
Notes: He1, RNase He1 from H. erinaceus; Po1, RNase Po1Citation5) from P. ostreatus; Ms, RNase MsCitation2) from A. saitoi; and T1, RNase T1Citation13) from A. oryzae. Numbers above the alignment correspond to the RNase He1 numbering. Residues in common with RNase He1 are shaded. ★: Catalytic site. The amino acid residues that were mutated are indicated by arrows. The site-directed mutants of RNase He1 are enclosed by dotted lines: 12RHe1, RNase He1 containing 12 mutations (12-mutant-He1); 4RHe1, RNase He1 containing four mutations (4-mutant-He1). The amino acid residues mutated into Asn or Gln in the two mutants are enclosed in the boxes.
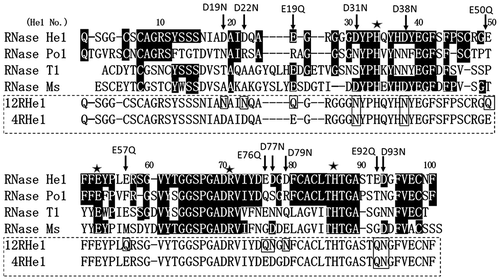
Table 1. Purification of 12RHe1 and 4RHe1.
Fig. 2. Tricine-SDS-PAGE of two mutant-He1s.
Notes: The details of the procedure are described in the text. a: Silver staining of molecular marker proteins. b: Silver staining of mutant-He1. (a), 12mutant-He1, (b), 4-mutant-He1.
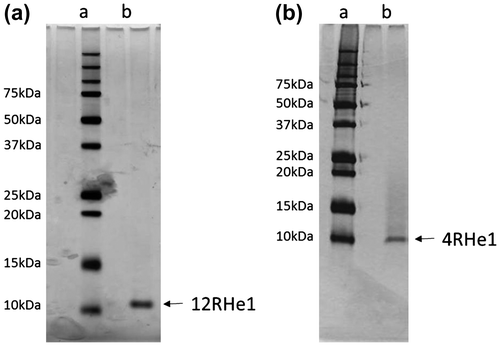
Fig. 3. Release of nucleotides upon digestion of RNA with RNase He1 and 12-mutant-He1.
The hydrolysis and separation conditions are described in the text. Symbols: -●-, 2′,3′ cGMP; ---●---, 3′GMP; -▲-, 2′,3′ cAMP + 3′AMP; -■-, 2′,3′ cCMP + 3′CMP; and -○-, 2′,3′ cUMP + 3′UMP.
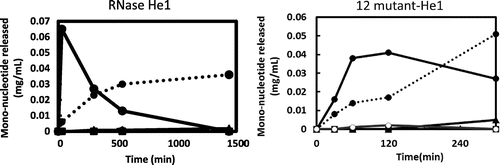
Fig. 4. Effect of the temperature and pH on the enzymatic activities of RNase He1, 12-mutant-He1, and 4-mutant-He1.
Notes: (a) Effect of the temperature on the enzymatic activities of RNase He1, 12-mutant-He1, and RNase T1. Enzyme activity was determined as described in the text using RNA as the substrate at various temperatures. The buffers (0.01 M) contained 1 mg/mL bovine serum albumin and 0.2 M NaCl. Activity was expressed as a percentage of the maximum activity. Symbols: ●, 12-mutant-He1; ○, RNase He1; and ▲, RNase T1. (b) Effect of pH on the enzymatic activity of RNase He1 and two mutants of RNase He1 (i.e. 4mutant-He1 and 12mutant-He1). Enzyme activity was determined as described in the text using RNA as the substrate. The buffers (0.01 M) used were acetate-NaOH buffer for pH 5.5–6.0 and Tris–HCl buffer for pH 6.5–8.5. Activity was expressed as a percentage of the maximum activity. Symbols: ●,4-mutant-He1; ▲, 12-mutant-He1; and ○, RNase He1.
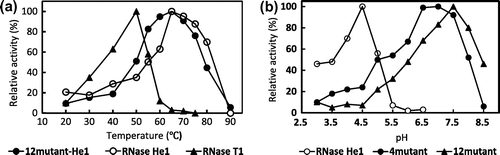
Fig. 5. Effects of 12mutant-He1, 4mutant-He1, and RNase He1 on the proliferation of HL-60 and Jurkat cells as determined with the MTT Assay.
Notes: Each point is the mean of three replicates and is reported as the percentage of the control, which lacked RNase. Cells were treated with a given concentration of RNases for 72 h. Cell proliferation without RNase was normalized to 100%. Symbols: ▲, RNase Po1; ●, 12mutant-He1; ○, RNase He1; ■, and 4mutant-He1.
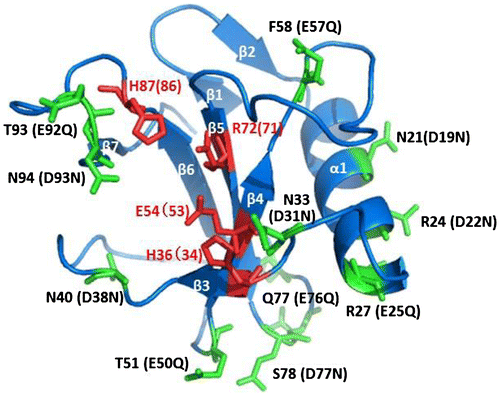
Fig. 6. The structural model of 12mutant-He1.
Notes: The location of the site-directed mutants of RNase He1 by structural modeling with the crystal structure of RNase Po1 as a template (PDB ID: 3WHO). The figure was drawn with PyMOL (http://pymol.sourceforge.net). α-Helices and β-strands are marked α1 and β1–7, respectively. Active site residues of RNase He1 are colored in red. The locations of the mutated amino acid residues in RNase He1 are colored in green. The amino acid numbers of RNase Po1 are shown followed by those of RNase He1 in parentheses.