Figures & data
Fig. 1. Multiple sequence alignment of the active site region of MtAPA and HIT superfamily proteins (S. cerevisiae Ap4A phosphorylases 1 and 2; APA1_Yeast and APA2_Yeast, Schizosaccharomyces pombe HIT family ApnA hydrolase; APH1_Schpo, H. sapiens HIT family ApnA hydrolase; Fhit_Human, Arabidopsis thaliana galactose-1-phosphate uridylyltransferases; GalT_Arath, and Oryctolagus cuniculus AMP-lysine hydrolase; Hint1_Rabbit).
Notes: Sequence alignment was performed using the ClustalW 2.1 software.Citation12) The HIT motif is underlined. The numbers along the top refer to the positions of amino acids in the sequence of MtAPA. The Ala-149 in MtAPA and the corresponding positions in other HIT superfamily proteins are shown in the box. In addition to Ala-149, the amino acids corresponding to Asn-139, Gly-146, Ser-147, His-153, and His-155 in MtAPA are shown in black, since these amino acids were previously presented as important residues for enzymatic activity of MtAPA,Citation2) and other amino acids are gray. The secondary structure of the active site based on the crystal structure of MtAPA is shown. The SWISS-PROT or TrEMBL accession numbers are as follows: MtAPA (P9WMK9), APA1_Yeast (P16550), APA2_Yeast (P22108), APH1_SCHPO (P49776), Fhit_Human (P49789), Galt_Arath (Q9FK51), and Hint1_Rabbit (P80912).
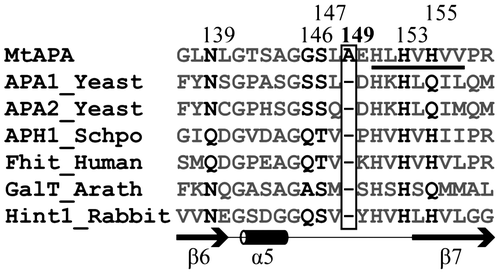
Table 1. Nucleotide substrate utilization by wild-type MtAPA and ∆149A-MtAPA.
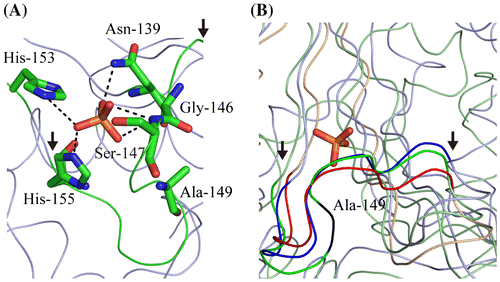
Fig. 2. Ribbon model of the active site of MtAPA (A) and the comparison of the loop conformation within it (B).
Notes: Ribbon diagrams were generated by PyMOL (W.L. DeLano, http://www.pymol.org). The atomic coordinates of crystal structures were downloaded from PDB (www.rcsb.org). It is likely that the phosphate ion-binding site of MtAPA (PDB ID; 3ANO) is the bona fide active site. In this site, Asn-139, Ser-147, His-153, and His-155 coordinate the phosphate ion, whereas Gly-146 is in close proximity.Citation2) (A) The loop in the active site is shown in green, with the start and end of the loop indicated by arrows. Asn-139, Gly-146, Ser-147, Ala-149, His-153, His-155, and the phosphate ion are represented by sticks. Carbon, nitrogen, oxygen, and phosphorus atoms are shown in green, blue, red, and orange, respectively. (B) The ribbon models of the active site of MtAPA, Fhit_Human (PDB ID; 6FIT), and GalT_Arath (PDB ID; 1Z84) are shown in green, blue, and red, respectively. The position of Ala-149 in MtAPA is shown in black. The phosphate ion in the active site of MtAPA is represented by sticks. The positions of the start and end of the loop in the active site are indicated by arrows.