Figures & data
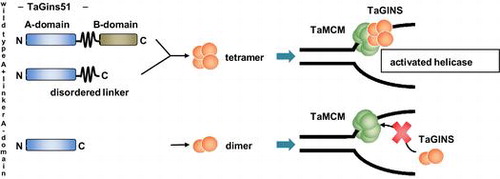
Table 1. Primers and oligonucleotides used in this study.
Fig. 1. Purification of TaGins51 proteins.
Notes: (A) Domain organization of the TaGins51 proteins used in this study. TaGins51-WT is composed of an α-helical A-domain, a β-stranded B-domain, and a disordered linker region between the two domains. (B) The purified TaGins51-WT, A + linker, and A-domain proteins (1 μg) were subjected to 10–20% SDS-PAGE, and were stained by Coomassie brilliant blue (left panel). For the Western blot analysis, 5 ng of the recombinant proteins were loaded onto the gel, followed by blotting and reactions with antibodies (right panel). M, molecular mass standards (BIO-RAD).
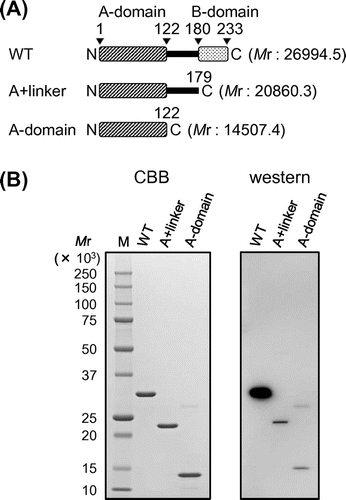
Fig. 2. Oligomeric state of TaGins51-mutants in solution.
Notes: A and B, Gel filtration analyses of the purified TaGins51-WT, A + linker, and A-domain proteins. The protein solutions were fractionated on a Superdex 200 3.2/30 (A) or Superdex 200 5/150 GL (B) column. The elution profiles monitored by the absorbance at 280 nm are shown. The oligomeric states of the eluted proteins were predicted, by calculating the molecular weights from the retention volumes in the chromatography.
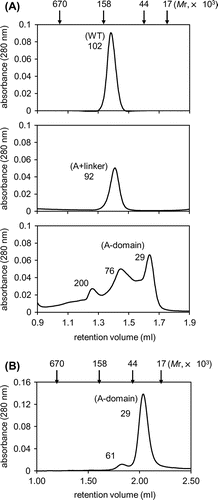
Fig. 3. Physical interaction between TaMCM and TaGins51-mutants.
Notes: (A) The DNA-binding activities of TaMCM, TaGins51-WT, A + linker, and A-domain are shown. The proteins were incubated with a 54 nt 32P-labeled ssDNA at 59 °C for 10 min, crosslinked with glutaraldehyde, and analyzed by 4% PAGE. The amounts of the proteins are shown at the top of each lane. (B) Antibody supershift assays were performed using the proteins shown at the top of each lane. Following crosslinking with glutaraldehyde, the reaction mixtures were incubated with either anti-TaMcm (M), anti-TaGins51 (G), or pre-immune (P) serum. The reaction products were analyzed by 4% PAGE.
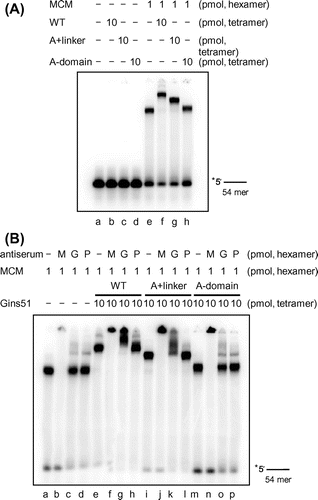
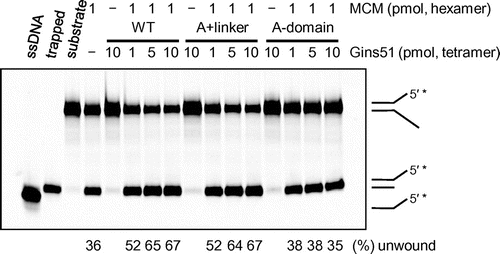
Fig. 4. The helicase activity of TaMCM in the presence of TaGins51-mutants.
Notes: The DyLight 800-labeled splayed-arm DNA substrate (0.5 pmol) was incubated with TaMCM and TaGins51-WT, A + linker, or A-domain, as described in the Materials and methods. The helicase activity is indicated at the bottom of the panel, as the relative amount of unwound DNA (%). Single-stranded DNA and trapped DNA were loaded in parallel, as controls for the unwinding reaction.