Figures & data
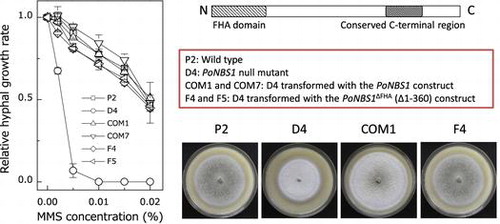
Table 1. Wild type and transgenic mutant strains of P. oryzae used in this study.
Table 2. Polymerase chain reaction primers used in this study.
Fig. 1. PoNBS1 contains conserved domains that are characteristic of the NBS1 protein.
Notes: (A) Domain structure of the typical NBS1 protein. The FHA domain is believed to be a phospho-specific protein–protein interaction motif. NBS1 binds to an MRN complex, ATM and ATR, which are proteins involved in DNA repair, at the conserved C-terminal region; (B) Amino acid alignment of the FHA domain from PoNBS1 and previously characterized NBS1 proteins. The sequences were aligned using CLUSTALW2 and processed with Boxshade. The residues highlighted in black boxes indicate the conserved amino acids. Sp, Schizosaccharomyces pombe; Sc, Saccharomyces cerevisiae; Hs, Homo sapiens; Mm, Mus musculus; and Dm, Drosophila melanogaster.

Fig. 2. Generation of PoNBS1 mutants.
Notes: (A) Schematic diagrams showing the strategy used for gene replacement in the PoNBS1 loci of the P. oryzae genome. The pDB51 construct was generated via ligation of the HPH-C, the amplified FL (upstream flanking sequence of PoNBS1) fragment and FR (downstream flanking sequence of PoNBS1) fragment to facilitate targeted gene deletion using the gene replacement approach. The restriction enzyme sites are abbreviated as follows: B, BamHI; S, SphI. (B) Confirmation of PoNBS1 disruption by Southern blot analysis. Genomic DNA extracted from the wild type (P2) and the PoNBS1-disrupted mutant (D4). DNA was digested with either BamHI (left) or SphI (right). The blots were probed with the FL (left) and HPH-C (right) fragments shown in (A). The results showed that the replacement of PoNBS1 with the knockout construct carrying HPH-C was accomplished successfully. M: position of the Lambda-HindIII DNA size standard. (C) The arrows indicate the primer positions for the genomic PCR. (D) Genomic PCR of the wild type and PoNBS1 mutants using the primer pair GP3up/GB513down. The 3.0-kb band corresponds to the genomic PoNBS1 gene and the 2.7-kb band corresponds to the PoNBS1 cDNA. M: position of the Lambda-HindIII DNA size standard. (E) Genomic PCR of the wild type and PoNBS1 mutants using the primer pair B51LOFHAup/GB513down. The 2.4-kb band corresponds to the PoNBS1△FHA cDNA. M: position of the Lambda-HindIII DNA size standard.
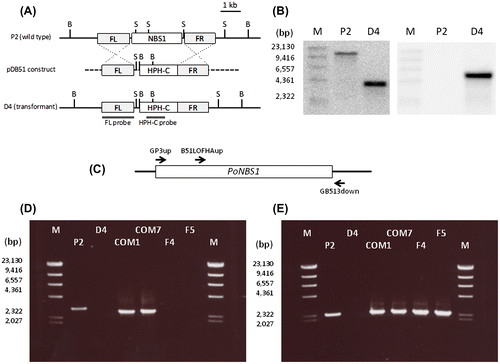
Fig. 3. Comparison of the sensitivity of the wild type and PoNBS1 mutants to mutagenic agents. The P. oryzae strains were cultured for eight days on YGA plates containing different concentrations of DNA damage-inducing agents, as indicated. Each point represents the mean ± SE (n ≥ 3).
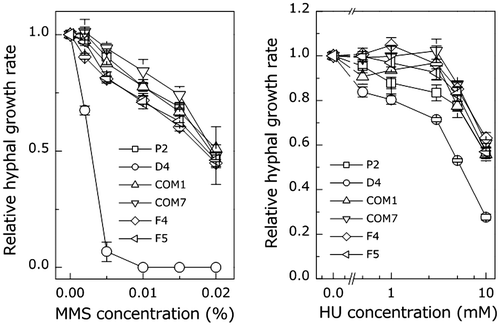
Fig. 4. Comparison of vegetative growth by the wild type and PoNBS1 mutants.
Notes: (A) Colony morphology of the wild type and PoNBS1 mutants. The P. oryzae strains were cultured on OMA plates for 13 days. Scale bar = 1 cm. (B) Quantification of hyphal growth by the wild type and PoNBS1 mutants grown on OMA plates for 13 days, as shown in (A). Each data point represents the mean ± SE (n ≥ 3).
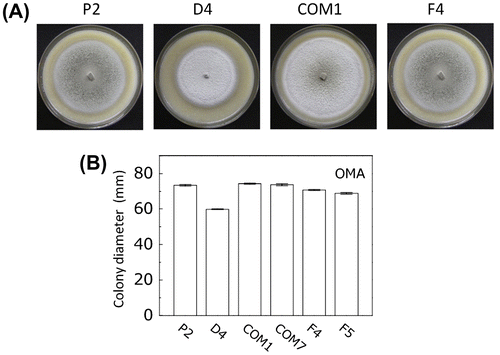
Fig. 5. Comparison of the conidial morphology in the wild type and PoNBS1 mutants. The numbers of septa were counted within the conidia of wild type and PoNBS1 mutants. Each data point represents the mean ± SE (n ≥ 100).
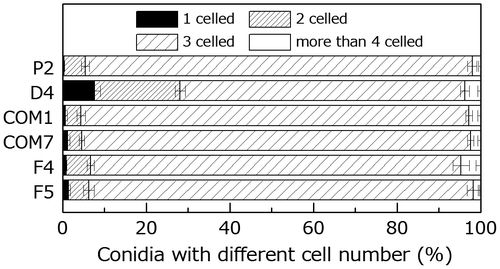
Fig. 6. Comparison of germination by the wild type and PoNBS1 mutants. The conidia from each strain were incubated at 25 °C for 6 or 24 h after inoculation. Each data point represents the mean ± SE (n ≥ 100).
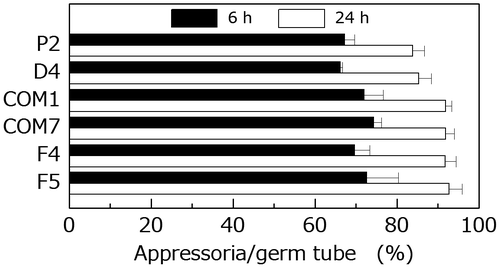
Fig. 7. Comparison of the frequency of appressorium formation by the wild type and PoNBS1 mutants. The conidia from each strain were incubated at 25 °C for 6 or 24 h after inoculation. Each data point represents the mean ± SE (n ≥ 100).