Figures & data
Fig. 1. Screening of candidate DH31R.
Notes: (A) A phylogenetic tree was generated based on the amino acid sequences of the following receptors: Drosophila DH31R, DH44R, and PDFR; Rhodnius DH31R-1B and DH31R-2B; Bombyx DHR and DH31R candidates (BNGR-B1, B2, B3, and B4). Phylogenetic analysis was performed with the neighbor-joining method using the ClustalX multiple alignment program and a bootstrap value of 1000 trials for each branch position. The indicated numbers are the bootstrap values as a percentage of 1000 replicates, and the scale bar indicates 0.05 changes per residue. Bootstrap values greater than 50% are indicated. The Mus musculus CR was used as an outgroup. (B) DH31-binding analysis of DH31R candidate receptors by examining the changes in the intracellular cAMP levels. BNGR-B1, B3, or B4-expressing HEK293 cells were treated with 1 μM DH31 and 0.5 mM IBMX. Each datum point represents the mean ± SEM (n = 3). Statistically significant differences were evaluated by Student’s t-test (***p < 0.001). (C) The dose response of DH31 was evaluated by the change in the intracellular cAMP levels in the BNGR-B1-expressed HEK293 cells. The cells were treated with 10−12 to 10−5 M of DH31 and 0.5 mM IBMX. Each datum point represents the mean ± SEM (n = 3).
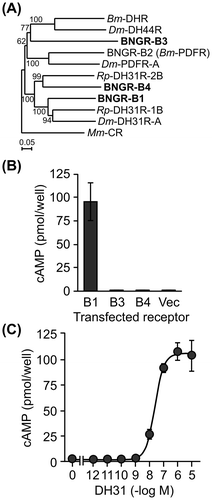
Fig. 2. Gene expression analysis of DH31 and BNGR-B1.
Notes: (A) The tissue distributions of DH31 and BNGR-B1 were evaluated by standard RT-PCR in the selected tissues of G0 larvae. BR: brain, SA: salivary gland, PG: prothoracic gland, ASG: anterior silk gland, FB: fat body, MT: Malpighian tubules, EP: epidermis, MG: midgut, TE: testis, and OV: ovary. (B) The distribution of DH31 was evaluated by standard RT-PCR in the CNS of G0 larvae. BR: brain, SOG: suboesophageal ganglion, TG1-3: thorax ganglion 1-3, AG1-8: abdominal ganglion 1-8. ((A) and (B)) Ribosomal protein L3 (RpL3) was used as an internal standard. The PCR primer sets used for the expression analysis were as follows: DH31: Fw, GTGCTGTGCTCCTGATCGTC, Rv, TGCGTTCCATCTGAATGAGG; BNGR-B1: Fw, TTCAACAATCGGACCTTGCC, Rv, GGTTCACGTCATCCTCCTCG; and RpL3: Fw, AGCACCCCGTCATGGGTCTA, Rv, TGCGTCCAAGCTCATCCTGC.
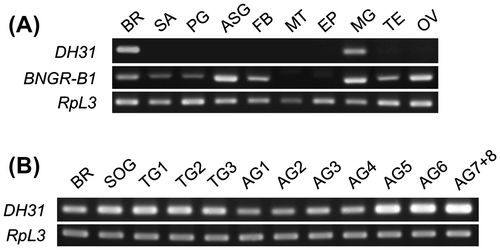
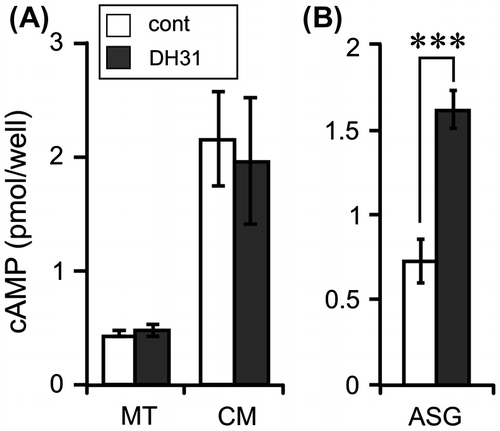
Fig. 3. Effects of DH31 on the cAMP levels in the Malpighian tubules and anterior silk gland.
Notes: (A) MT (approx. 1 cm long) was cultured in 100 μL of Grace’s insect culture medium (Gibco, Grand Island, NY) with 1 μM DH31 and 0.5 mM IBMX at 25 °C for 1 h. The amount of cAMP was measured in the MT and cultured medium (CM). Each datum point represents the mean ± SEM (n = 8). (B) ASG (approx. 2.5 cm long) was cultured in 300 μL of Grace’s insect culture medium with 1 μM DH31 and 0.5 mM IBMX at 25 °C for 30 min. Each datum point represents the mean ± SEM (n = 6). cAMP was extracted from the tissues by 100 μL of acidic ethanol (0.1% 10 N HCl, v/v), and the supernatant was used after evaporation. Statistically significant differences were evaluated by Student’s t-test (***p < 0.001).