Figures & data
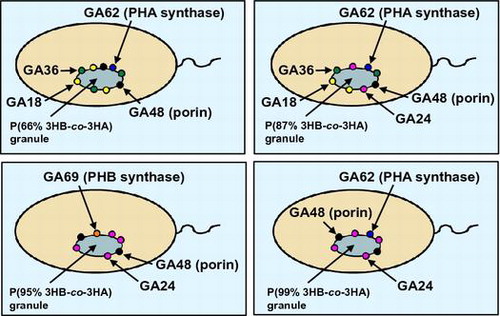
Fig. 1. Organization of pha and phb loci in Pseudomonas sp. 61-3.
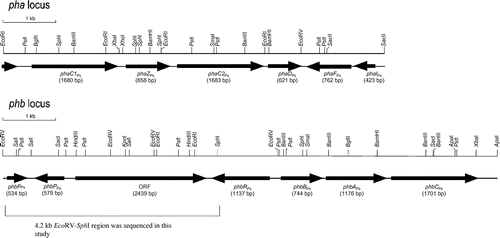
Table 1. Bacterial strains and plasmids used in this study.
Fig. 2. SDS-PAGE analysis of native PHA granules isolated from the recombinant strains of Pseudomonas sp. 61–3. Lane 1, molecular weight markers; lane 2, Pseudomonas sp. 61–3 (phbC::tet); lane 3, Pseudomonas sp. 61–3 (phbC::tet)/pJASc22; lane 4, Pseudomonas sp. 61–3 (phbC::tet)/pJKSc46-pha; lane 5, Pseudomonas sp. 61–3 (phbC::tet)/pJKSc54-phab; lane 6, Pseudomonas sp. AC1-TnK; and lane 7, Pseudomonas sp. BCG-TcGm/pJKSc54-phab.
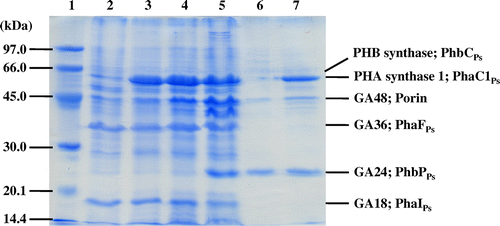
Table 2. Relationship of the monomer composition of PHA accumulated by recombinant strains of Pseudomonas sp. 61-3 and the granule-associated proteins.
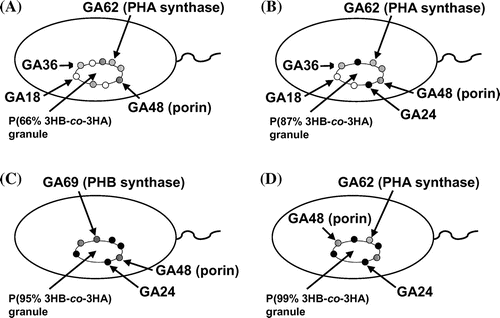
Fig. 3. The localization model of the proteins associated with polyester granules accumulated in (A) Pseudomonas sp. 61–3 (phbC::tet)/pJKSc46-pha, (B) Pseudomonas sp. 61–3 (phbC::tet)/pJKSc54-phab, (C) Pseudomonas sp. AC1-TnK, and (D) Pseudomonas sp. BCG-TcGm/pJKSc54-phab.