Figures & data
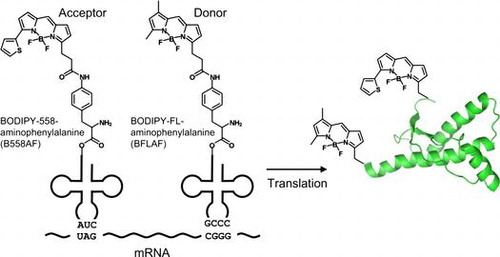
Fig. 1. Schema of double-fluorescent labeled PrP synthesis.
Fig. 2. The introduction of mutations into expression vector.

Table 1. Concentrations of double-fluorescent labeled PrP synthesized in this study.
Fig. 3. Analysis of purified, double-labeled protein containing BFLAF and B558AF.
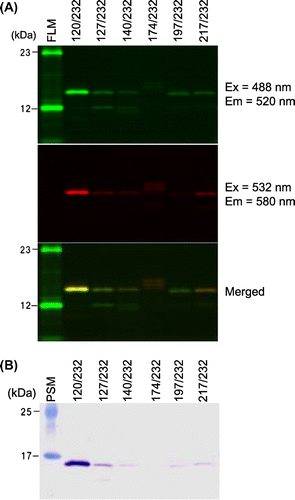
Fig. 4. Each double-labeled protein 140/232 from the original and optimized sequences was separated into total fraction (pre-separation, T), supernatant fraction (S), and pellet fraction (P).
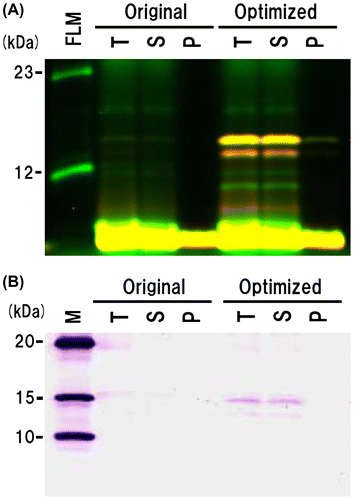
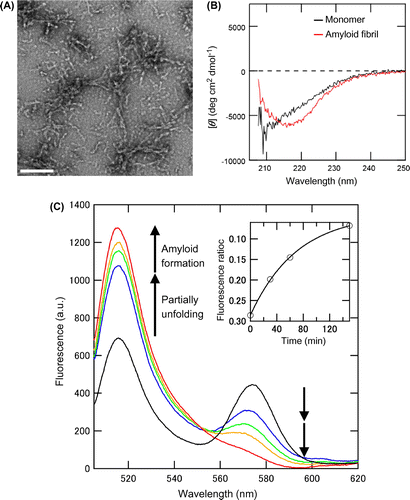
Fig. 5. Amyloid fibril formation of the double-labeled protein 120/232.