Figures & data
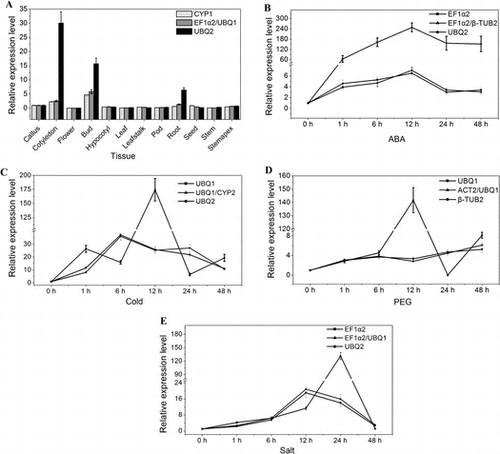
Table 1. Details of primers and parameters derived from qRT-PCR in this study.
Fig. 1. Cycle threshold (Ct) values of the candidate reference genes across the experimental samples.
Notes: The box represented the 25th and 75th percentiles of data. Whiskers represented the maximum and minimum values. Horizontal line within the box represented the median Ct value.
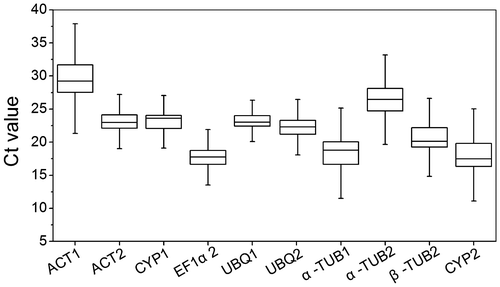
Fig. 2. Average expression stability (M) values of candidate reference genes by geNorm.
Notes: (A) The M values calculated for all plant samples. (B) The M values calculated for 12 various tissues. (C) The M values calculated under ABA treatment. (D) The M values calculated under cold stress. (E) The M values calculated under PEG treatment. (F) The M values calculated under salt stress.
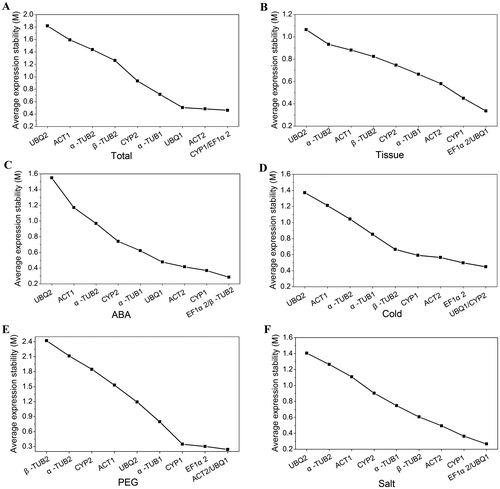
Fig. 3. Pairwise variation analysis of the candidate reference genes.
Notes: Pairwise variation (Vn/Vn+1) was analyzed between the normalization factors NFn and NFn+1. Arrows indicated the optimal number of reference genes required for normalization.
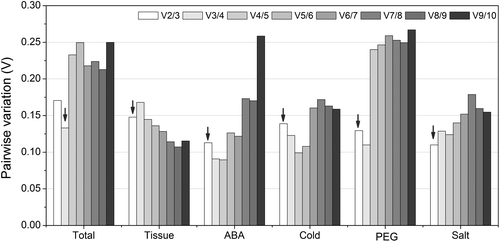
Table 2. Ranking of the candidate reference genes according to their stability estimated by NormFinder.
Table 3. Ranking of the candidate reference genes according to their stability calculated by BestKeeper.
Table 4. Ranking of candidate reference gene stability according to geNorm, NormFinder, and BestKeeper.
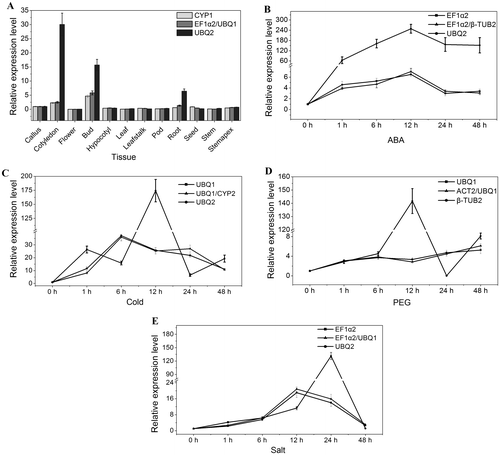
Fig. 4. The relative transcript level of WRKY gene in 12 tissues and under different stress normalized by the selected reference genes.
Notes: (A) The relative transcript level of WRKY gene in 12 tissues. (B) The relative transcript level of WRKY gene under ABA treatment. (C) The relative transcript level of WRKY gene under cold stress. (D) The relative transcript level of WRKY gene under PEG treatment. (E) The relative transcript level of WRKY gene under salt stress.