Figures & data
Fig. 1. Multiple alignments of the amino acid sequences of 3FTxs from various species and the structure model of the α/γ subunit interface interacting with Pr-SLTX.
Notes: (A) We isolated the cDNAs of 3FTxs from the venom gland cDNA library of P. rossignolii. The amino acid sequences of three-finger toxins from Elapid snakes were aligned using ClustalW program. The protein accession numbers are the following: P. australis SNTX, ABK63527; Oxyuranus scutellatus SNTX-1, AAZ22673; P. textilis SNTX, ABK63535; Erabutoxin a, BAA75752; P. australis LNTX, ABK 63539; Austrelaps labialis LNTX 23B, ABX58159; α-bungarotoxin, AAL30056; α-bungarotoxin, CAA69971; O. scutellatus LNTX-1, ABK63543; and P. textilis LNTX, ABK63540. Both O. scutellatus LNTX-1 and P. textilis LNTX have four disulfide bonds, and they did not follow the nomenclature of short- and long-type 3FTs. Residues that are identical among the majority of the amino acid sequences are highlighted in gray, and conserved cysteine residues are highlighted in black. The number of amino acid residues is given to the right of each line in italics. Intervals of 10 amino acids are indicated by dots above the alignment. Gaps (-) have been inserted to achieve maximum homology. The nucleotide sequence of Pr-SNTX has been assigned DDBJ/EMBL/GenBank Accession Number AB778565. (B) Overall view of the structure model from the side for the α/γ subunit interface interacting with Pr-SLTX. (C) Overall view of the structure model from the top. The three-dimensional structure of Pr-SNTX was modeled using MODELER v. 9.11 with the structure of cobrotoxin (PDB 1V6P) as a template. Pr-SLTX, α-subunit, and γ-subunit are displayed in magenta, green, and cyan, respectively. Side chains of amino acid residues (Lys27, Trp29, Asp31, His32, Arg33, and Lys47) important for toxin binding to nAChR Citation13) are shown in blue.
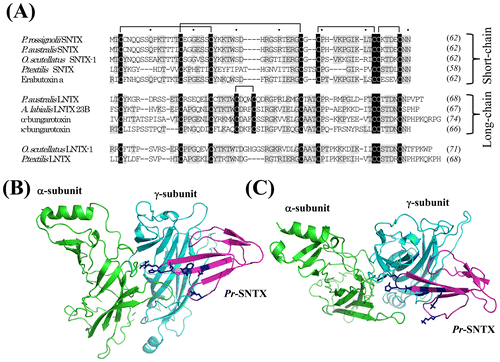
Fig. 2. SDS-PAGE and MALDI-TOF-MS spectrometry analyses of recombinant Pr-SNTX protein.
Notes: (A) Recombinant Pr-SLTX protein was expressed in E. coli and purified by TALON affinity column and a reverse-phase HPLC column, Intrada WP-RP. Purified protein (2 μg) was loaded on 12% NuPAGE (Life Technologies, Carlsbad, CA) and stained with Coomassie Brilliant Blue R-250. (B) The molecular weight of recombinant Pr-SNTX was verified by MALDI-TOF-MS using AXIMA Performance. The observed average molecular mass [M + H]+ of recombinant Pr-SNTX protein was 6949.66. The mass peaks at m/z 3476.14 correspond to [M + 2H]2+ for Pr-SNTX.
![Fig. 2. SDS-PAGE and MALDI-TOF-MS spectrometry analyses of recombinant Pr-SNTX protein.Notes: (A) Recombinant Pr-SLTX protein was expressed in E. coli and purified by TALON affinity column and a reverse-phase HPLC column, Intrada WP-RP. Purified protein (2 μg) was loaded on 12% NuPAGE (Life Technologies, Carlsbad, CA) and stained with Coomassie Brilliant Blue R-250. (B) The molecular weight of recombinant Pr-SNTX was verified by MALDI-TOF-MS using AXIMA Performance. The observed average molecular mass [M + H]+ of recombinant Pr-SNTX protein was 6949.66. The mass peaks at m/z 3476.14 correspond to [M + 2H]2+ for Pr-SNTX.](/cms/asset/e862a90a-673f-4665-94fd-5eb699621968/tbbb_a_1065169_f0002_b.gif)
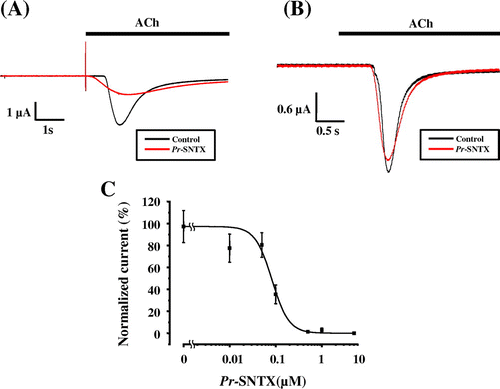
Fig. 3. Pr-SNTX inhibits the acetylcholine-evoked current in oocyte expressed muscle-type nAChR (α2βδε), but not neuronal α7 nAChR.
Notes: (A) Acetylcholine-evoked currents of muscle-type nAChRs (α2βδε) were recorded in the absence (black) or presence (red) of 100 nM Pr-SNTX protein; bar indicates acetylcholine (10 μM) application. (B) Acetylcholine-evoked currents of neuronal α7 nAChRs were recorded in the absence (black) or presence (red) of 10 μM Pr-SNTX protein; bar indicates acetylcholine (100 μM) application. (C) Dose-dependent inhibitory effects of Pr-SNTX. Data were analyzed and plotted using Origin 8 (OriginLab., Northampton, MA, USA). The 100% activity value was determined by measurements in the absence of Pr-SNTX protein. Each data point represents the mean from a single experiment performed in triplicate. Mean values obtained from five oocytes. Error bars show the standard error of the mean.