Figures & data
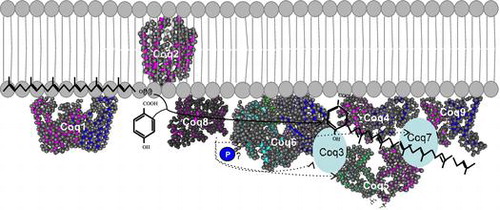
Fig. 1. The electron transfer system and enzymes involved in CoQ biosynthesis. Notes: The position of CoQ in the electron transfer system is shown. Complexes I and II transfer electrons to CoQ from NADH and FADH2, respectively. In yeast, NADH dehydrogenase replaces Complex I in the first reaction. Electrons are transferred to Complex III from CoQH2, a reduced form of CoQ, and then further transferred to Complex IV through cytochrome c (Cytc). Protons are transferred to the intermembrane space, and this proton gradient drives ATP production through Complex V. A number of different enzymes are coupled with CoQ in oxidation–reduction reactions: DHODH, dihydroorotate dehydrogenase; SQR, sulfide quinone reductase; ETFDH, electron transfer flavoprotein dehydrogenase; and GPDH, glycerol-3-phosphate dehydrogenase.
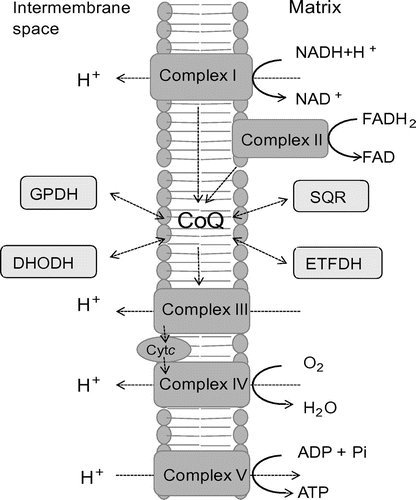
Fig. 2. Biosynthetic pathway of the isoprenoid tail of CoQ. Notes: Isopentenyl diphosphate (IPP) is synthesized via the mevalonate (MVA) pathway in eukaryotes and the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway in prokaryotes and plants. In each organism, trans-polyprenyl diphosphate of a certain length is synthesized by polyprenyl diphosphate synthase. S. cerevisiae Coq1 synthesizes six isoprene units, E. coli IspB synthesizes eight isoprene units, and H. sapiens or S. pombe decaprenyl diphosphate (DPP; a heteromer of PDSS1 and PDSS2 or Dps1 and Dlp1, respectively) synthesize ten isoprene units. S. cerevisiae Coq2, E. coli UbiA, and H. sapiens COQ2 or S. pombe Ppt1(Coq2) condense PHB with trans-polyprenyl diphosphate to form CoQ6, CoQ8, and CoQ10, respectively. DMAPP, dimethylallyl diphosphate; GPP, geranly diphosphate; FPP, farnesyl diphosphate; IPP, isopentenyl diphosphate; HexPP, Hexaprenyl diphosphate; OPP, Octaprenyl diphosphate.
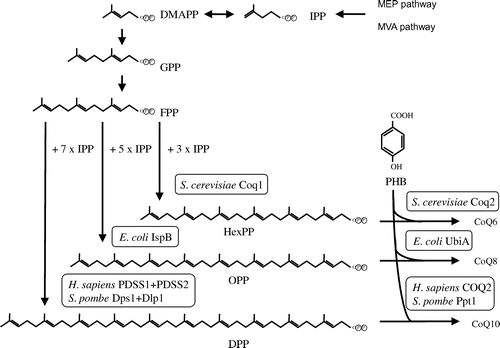
Fig. 3. Overview of the proposed CoQ biosynthetic pathway. Notes: The CoQ biosynthetic pathways of prokaryotes (represented by E. coli) and eukaryotes (represented by S. cerevisiae). In general, the gene names ubi* and COQ(coq)* are used in prokaryotes and eukaryotes, respectively; however, the nomenclature of genes can differ among species. After condensation of PHB with trans-polyprenyl diphosphate, the ring structure is modifed. In E. coli, decarboxylation by UbiD or UbiX is followed by hydroxyation by UbiI, O-methylation by UbiG, hydroxyation by UbiH, C-methylation by UbiE, a final hydroxylation by UbiF, and then O-methylation by UbiG. At least eight genes are responsible for CoQ biosynthesis in E. coli. In S. cerevisiae, pABA is also used as a substrate in addition to PHB. In this species, the first ring is modifed by hydroxyation by Coq6, followed by O-methylation by Coq3; however, the subsequent decarboxylation and hydroxylation steps are unclear. The ring is then modified further by C-methylation by Coq5, a final hydroxylation by Coq7, and O-methylation by Coq3.
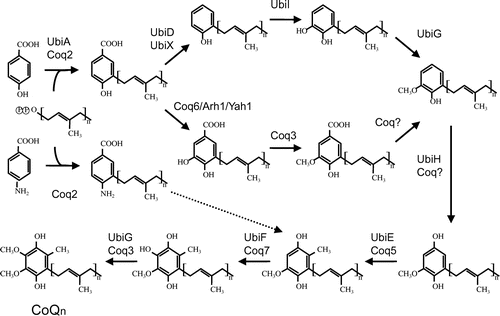
Table 1. CoQ biosynthetic genes from various species.
Table 2. Genotype–phenotype correlations in inherited deficiencies of CoQ10 biosynthesis in humans.
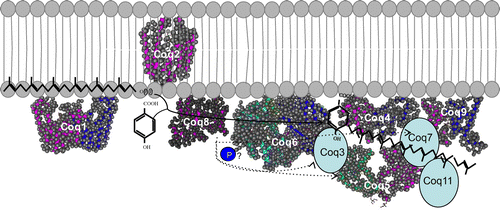
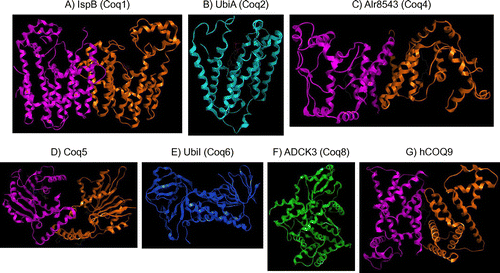
Fig. 5. Structure of the CoQ biosynthetic enzyme complex. Notes: The enzymes involved in CoQ synthesis form a complex in S. cerevisiae.Citation78) There is also some evidence that this complex exists in humans. This figure is modified from the figure reported by Allan et al.Citation72) Proteins in the figure are not proportional to the actual molecular sizes. The structure of S. cerevisiae Coq5 has been solved; for the other enzymes, the structures of homologs from other species are indicated. E. coli IspB is shown as a Coq1 homolog, A. pernix UbiA is shown as a Coq2 homolog, Alr8543 from Nostoc sp. PCC7120 is shown as a Coq4 homolog, E. coli UbiI is shown as a Coq6 homolog, human ADCK3 is shown as a Coq8 homolog, and human COQ9 is shown as a Coq9 homolog. Coq1 is separated from the complex. Coq2 spans the inner membrane. The positions of the other proteins have not been defined experimentally, although Coq4 seems to be in the center, and Coq7 and Coq8 seem to be located at the edge of the complex.