Figures & data
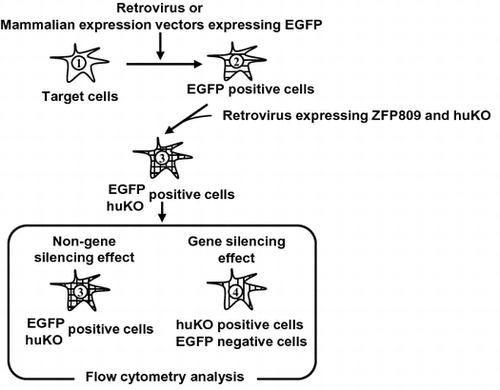
Fig. 1. Schematic diagram of fluorescent reporter-containing vector system combined with flow cytometry methodology.
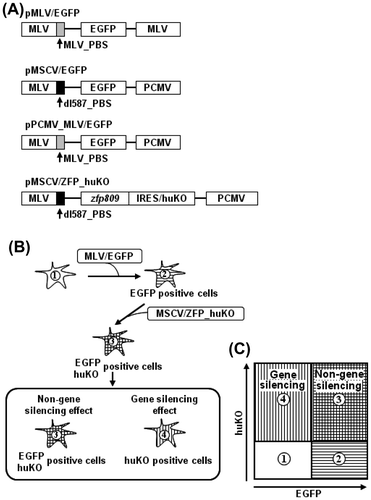
Fig. 2. Long-term monitoring of ZFP809-mediated gene silencing effect on transgene expression driven by MLV.
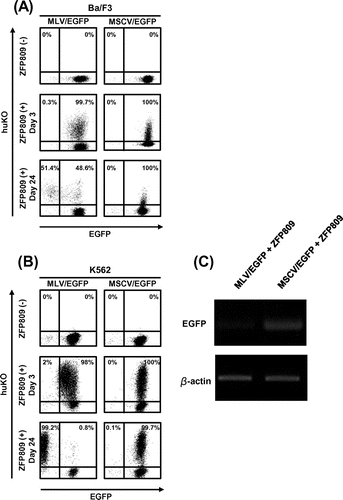
Fig. 3. Long-term monitoring of ZFP809-mediated gene silencing effect on transgene expression driven by CMV or EF1-α promoters.
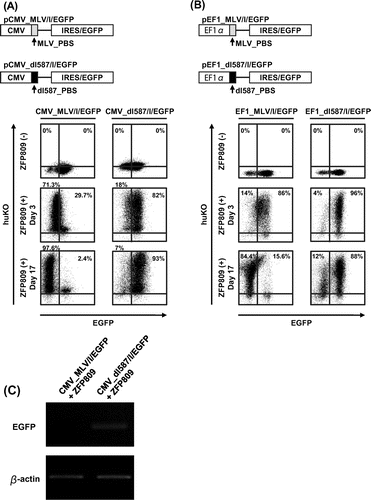
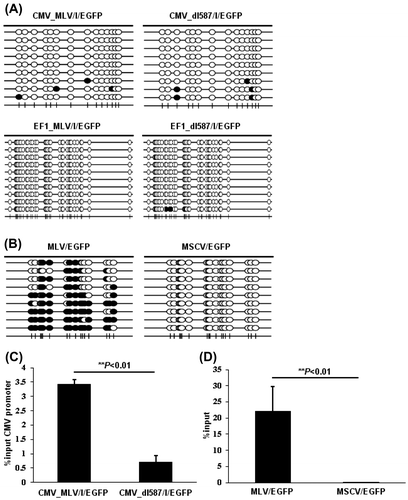
Fig. 4. Epigenetic modifications of CMV and EF1-α promoter induced by ZFP809.