Figures & data
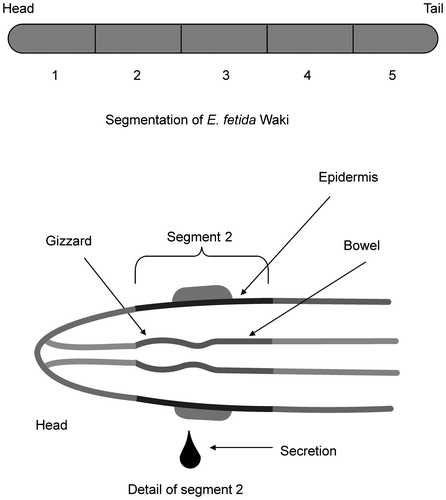
Table 1. Enzyme activity of crude extract from each segment of E. fetida Waki.
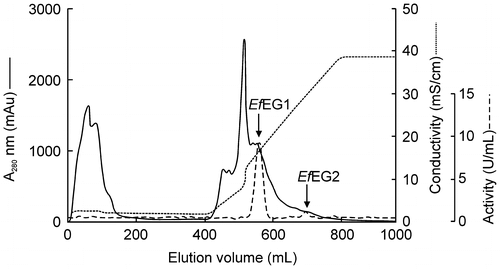
Table 2. Elution profile of EG isozymes in E. fetida Waki from a DEAE-Toyopearl column.
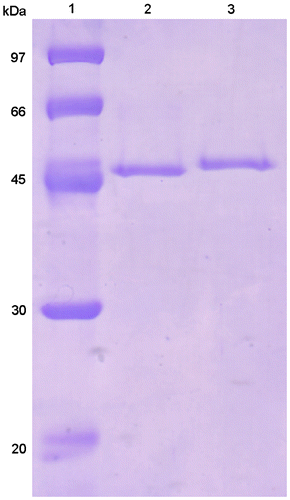
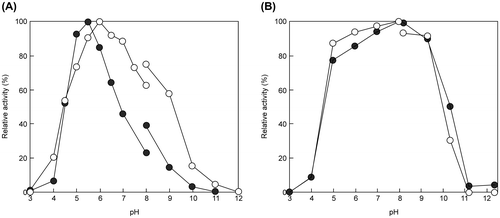
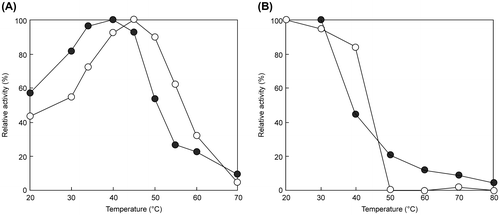
Table 3. Properties of EGs from animal and T. reesei QM9414.
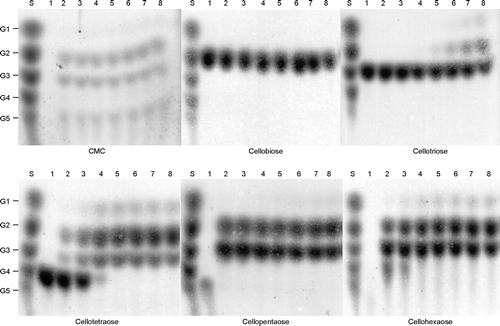
Table 4. Substrate specificity of purified EfEG1 and EfEG2 from E. fetida Waki.
Ueda M, Ito A, Nakazawa M, Miyatake K, Sakaguchi M, Inouye K. Cloning and expression of the cold-adapted endo-1,4-β-glucanase gene from Eisenia fetida. Carbohydr. Polym. 2014;101:511–516.10.1016/j.carbpol.2013.09.057 Van Arsdell JN, Kwok S, Schweickart VL, Ladner MB, Gelfand DH, Innis MA. Cloning, characterization, and expression in Saccharomyces cerevisiae of endoglucanase I from Trichoderma reesei. Bio/Technol. 1987;5:60–64.10.1038/nbt0187-60 Saloheimo M, Lehtovaara P, Penttilä M, et al. EGIII, a new endoglucanase from Trichoderma reesei: the characterization of both gene and enzyme. Gene. 1988;63:11–21.10.1016/0378-1119(88)90541-0 Nakazawa H, Okada K, Onodera T, Ogasawara W, Okada H, Morikawa Y. Directed evolution of endoglucanase III (Cel12A) from Trichoderma reesei. Appl. Microbiol. Biotechnol. 2009;83:649–657.10.1007/s00253-009-1901-3 Watanabe H, Nakamura M, Tokuda G, Yamaoka I, Scrivener AM, Noda H. Site of secretion and properties of endogenous endo-β-1,4-glucanase components from Reticulitermes speratus (Kolbe), a Japanese subterranean termite. Insect Biochem. Mol. Biol. 1997;27:305–313.10.1016/S0965-1748(97)00003-9 Suzuki K, Ojima T, Nishita K. Purification and cDNA cloning of a cellulase from abalone Haliotis discus hannai. Eur. J. Biochem. 2003;270:771–778.10.1046/j.1432-1033.2003.03443.x Kim N, Choo YM, Lee KS, et al. Molecular cloning and characterization of a glycosyl hydrolase family 9 cellulase distributed throughout the digestive tract of the cricket Teleogryllus emma. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2008;150:368–376.10.1016/j.cbpb.2008.04.005 Lee SJ, Kim SR, Yoon HJ, et al. cDNA cloning, expression, and enzymatic activity of a cellulase from the mulberry longicorn beetle, Apriona germari. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2004;139:107–116. Lee SJ, Lee KS, Kim SR, et al. A novel cellulase gene from the mulberry longicorn beetle, Apriona germari: gene structure, expression, and enzymatic activity. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2005;140:551–560.10.1016/j.cbpc.2004.12.003