Figures & data
Fig. 1. Transactivation of 5xGAL4-UAS::LUC reporter gene by GAL4-BD/CIGR2 in suspension-cultured rice cells.
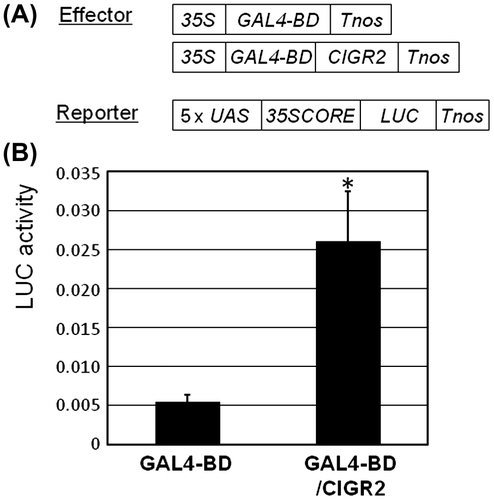
Fig. 2. Activation of OsHsf23 by CIGR2.
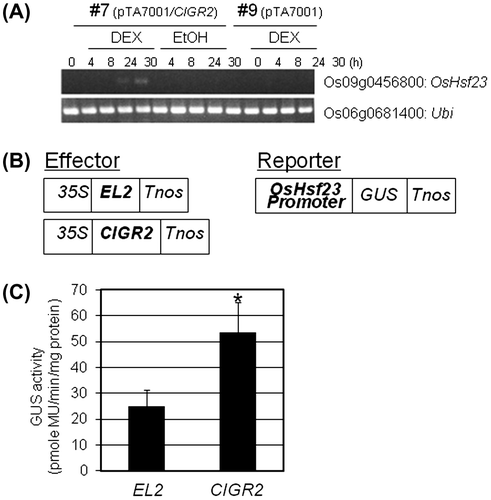
Fig. 3. Ratio of granulated cells in the epidermis of leaf sheath of CIGR2-RNAi and OsHsf23-RNAi lines inoculated with an avirulent isolate of M. oryzae, P91-15B.
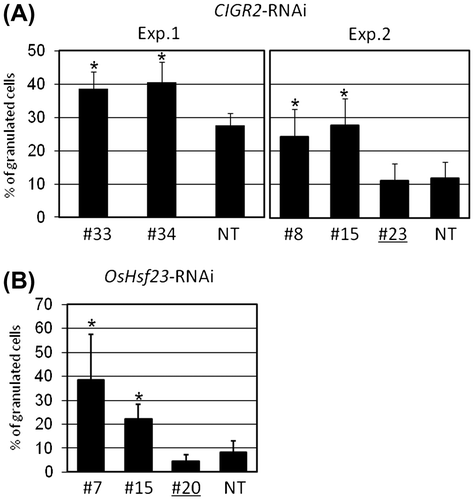
Fig. 4. Length of lesions formed on the leaves of CIGR2-RNAi lines.
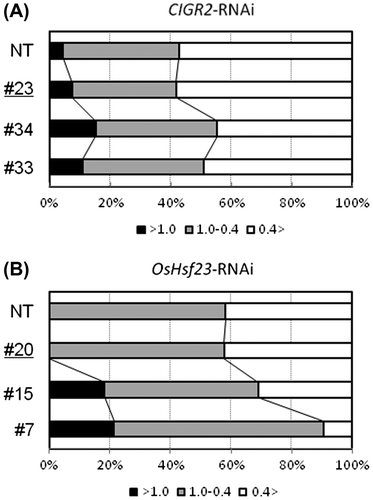
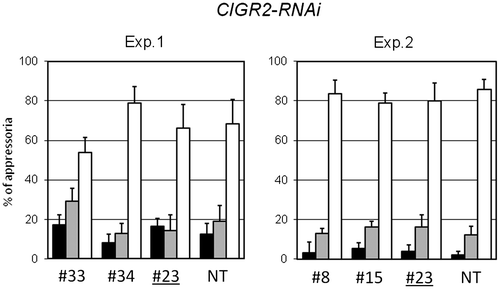
Fig. 5. Leaf sheath assay of CIGR2-RNAi lines.