Figures & data
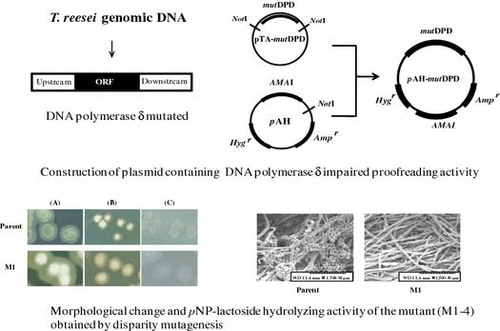
Fig. 1. Scheme for the construction of pAH and pAH-mutDPD.
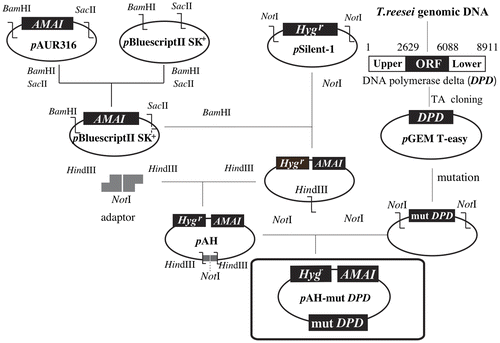
Fig. 2. Plasmid insertion confirmed by PCR.
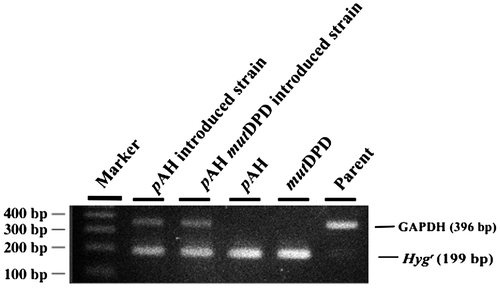
Table 1. Mutation ratio of disparity mutagenesis evaluated by colony formation in comparison to UV-irradiation.
Fig. 3. Colony formation of glucose de-repressed mutant and parent.
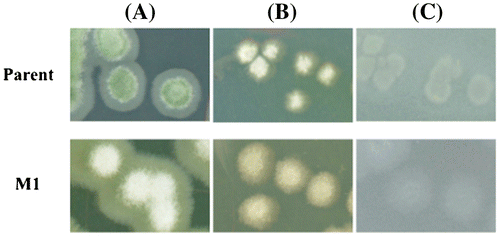
Fig. 4. Scanning electron microscopy of conidia and mycelia.
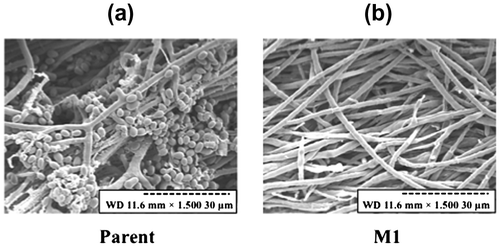
Fig. 5. Expression of cre1 analyzed by RT-PCR.
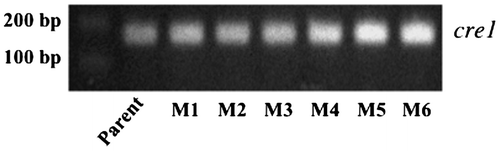
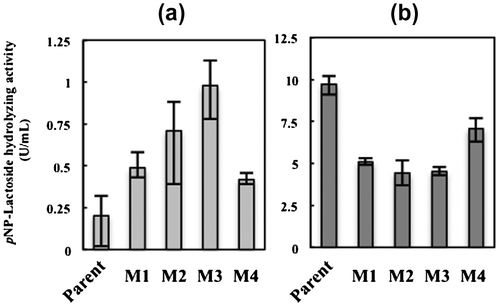
Fig. 6. Cellulase activity of the glucose de-repressed mutants (M1–4) and parent strain.