Figures & data
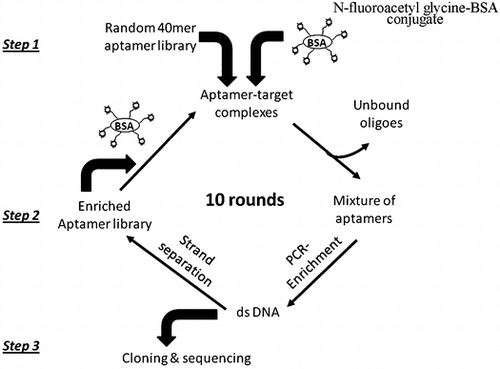
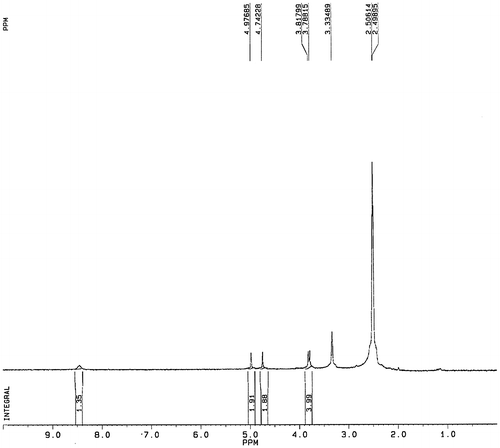
Fig. 1. Structure formula of fluoroacetamide and its analog, N-fluoroacetyl glycine (A) and synthetic steps of N-fluoroacetyl glycine (B).
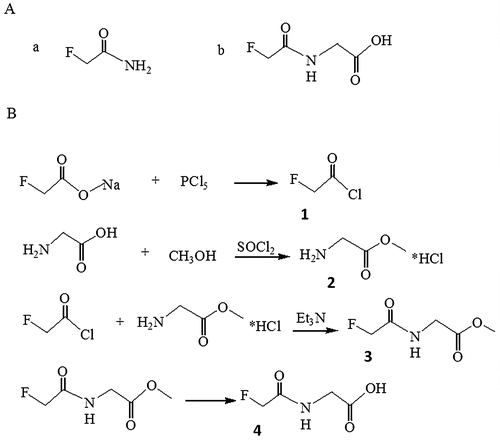
Table 1. Sequences of the 40nt core regions of the cloned aptamers.
Fig. 4. Analysis of PCR products from each round of SELEX enrichment. PCR products were sampled after each round of enrichment and run on a 2% agarose gel.
Notes: M: molecular ladder (50–500 bp), Lanes 1-10: PCR products from enrichment rounds 1-10, and lane 11: no template control.
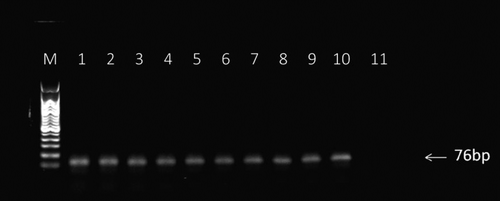
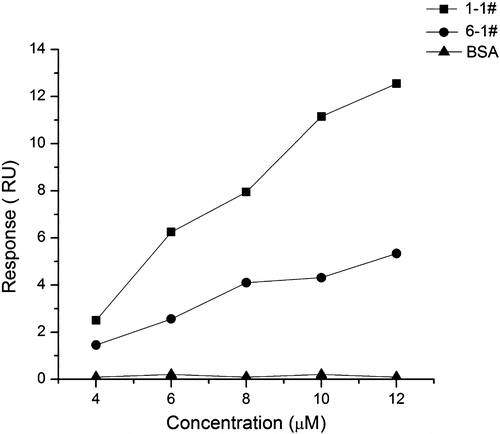
Fig. 5. Affinity of the selected ssDNA to the target in each selection round. The initial ssDNA library was used as reference.
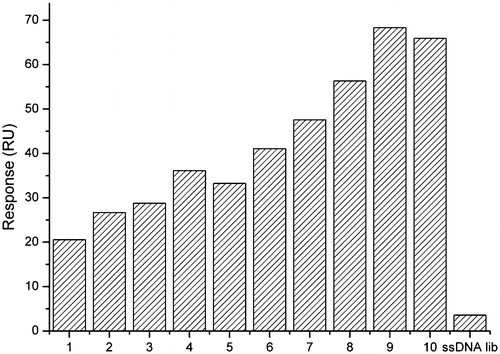
Fig. 6. Multiple sequence alignments of the selected aptamers (without primers).
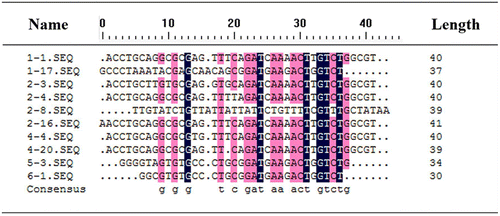
Fig. 7. Predicted secondary structures of aptamer 1-1, 6-1, 2-8, and 1-17.
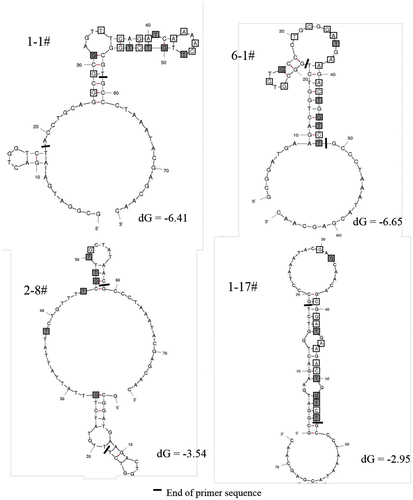
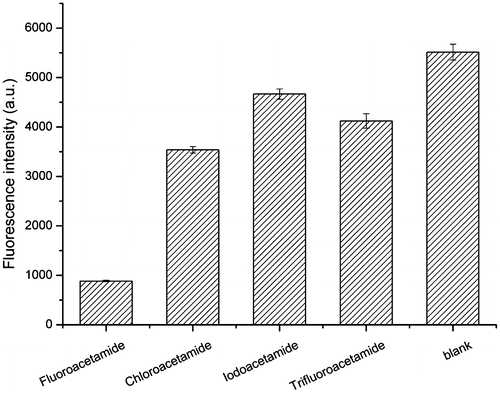
Fig. 10. Characterization of specificity of aptamer 1-1 for fluoroacetamide. The binding affinity of aptamer 1-1 to fluoroacetamide and other structurally similar molecules was tested by a dipstick assay based on lateral-flow technology. The fluorescence intensity at test line for each target was measured. The buffer solution without any target was used as blank.