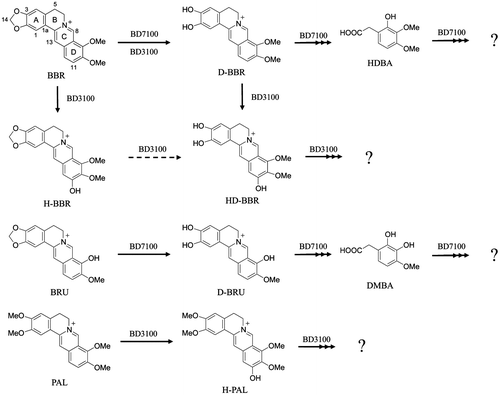
Figures & data
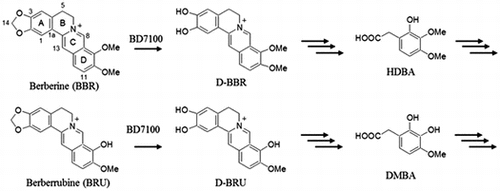
Fig. 1. Effect of initial medium conditions (temperature and pH) on the growth of BD7100.
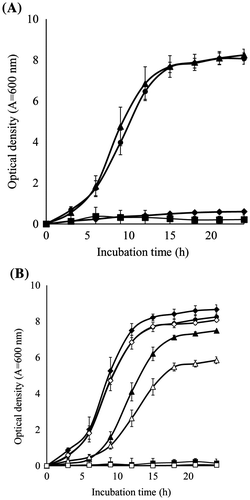
Fig. 2. The growth of BD7100 and BBR degradation in the growing-cell assay.
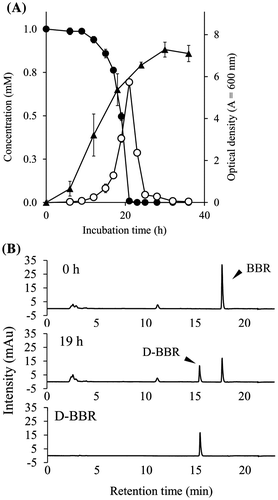
Fig. 3 The degradation of BBR by resting cells of BD7100 and the resulting metabolites.
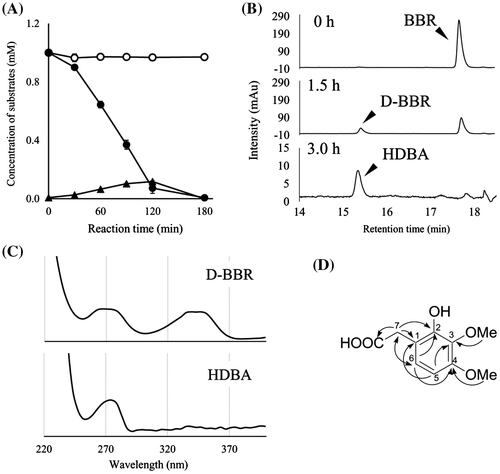
Fig. 4. The degradation of BRU by resting cells of BD7100 and the resulting metabolites.
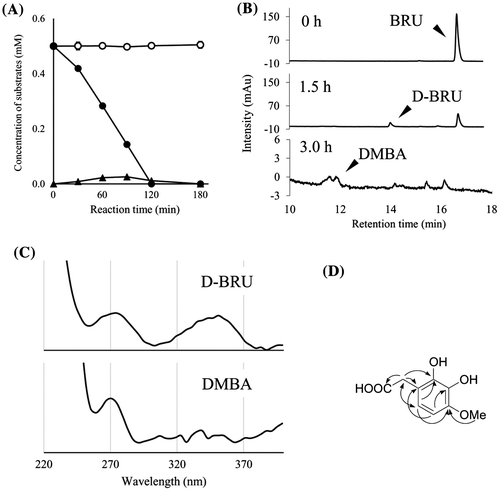
Fig. 5. The inferred pathways for degradation of BBR and its analogs in BBR-utilizing bacteria.