Figures & data

Table 1. The Neurospora crassa strains used in this study.
Fig. 1. Comparison of the growth and morphology of mak-1, mak-2, and os-2 disruptants of Neurospora crassa.
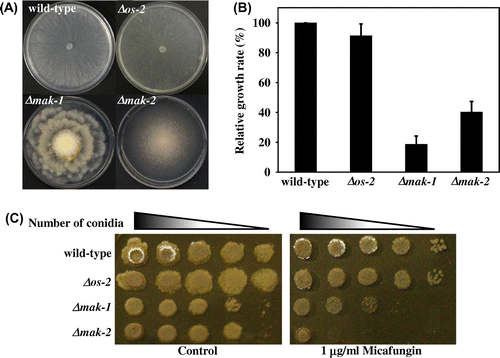
Fig. 2. Phosphorylation of MAK-1 and MAK-2 MAPKs is induced by micafungin.
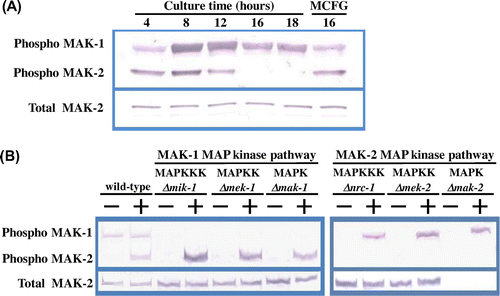
Table 2. Expression analysis of 12 cell wall biogenesis-related genes by quantitative real-time RT-PCR.
Table 3. Microarray analysis of genes upregulated by micafungin treatment in wild-type strain.
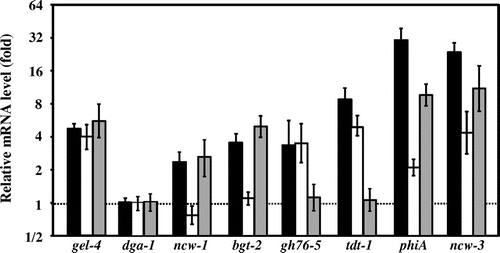
Fig. 3. Relative mRNA levels of selected genes in response to micafungin.