Figures & data
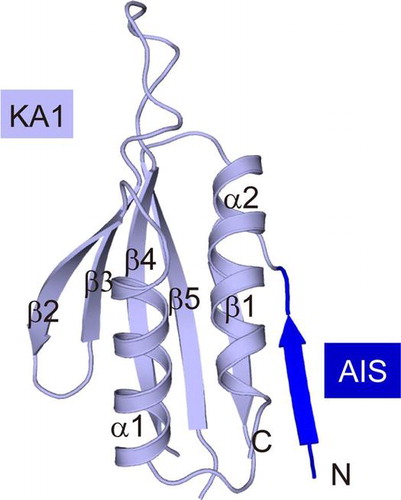
Fig. 1. Biochemical characteristics of SAD-B KD-UBA.
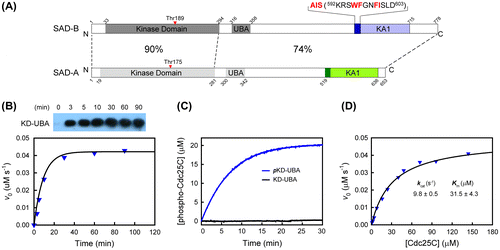
Table 1. Data collection and refinement statistics.
Fig. 2. Overall structure of AIS-KA1 of mouse SAD-B.
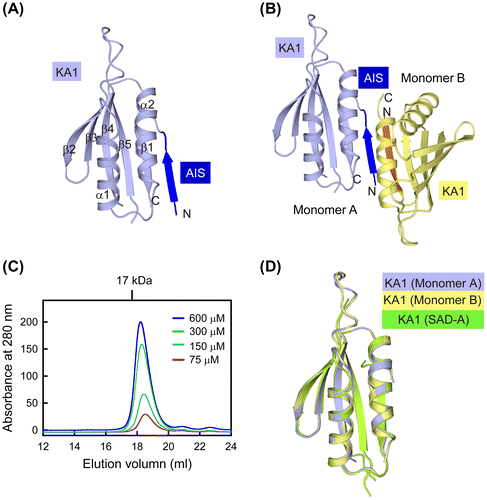
Fig. 3. Analysis of the AIS sequence in the free AIS-KA1 structure.
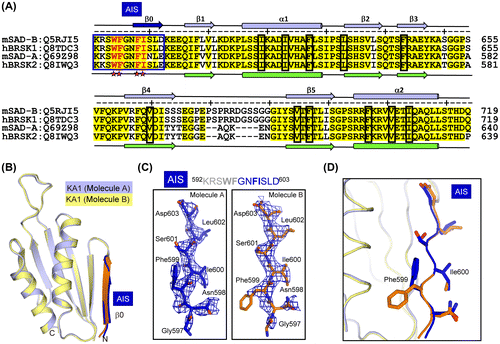
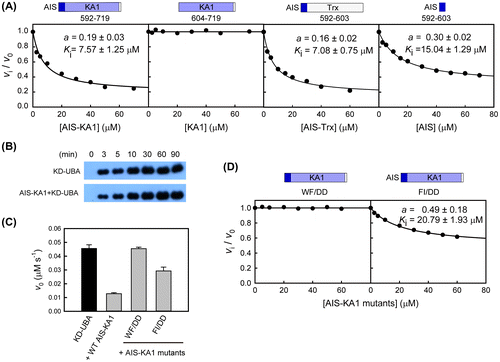
Fig. 4. The conserved AIS inhibits the kinase activity of SAD-B.