Figures & data
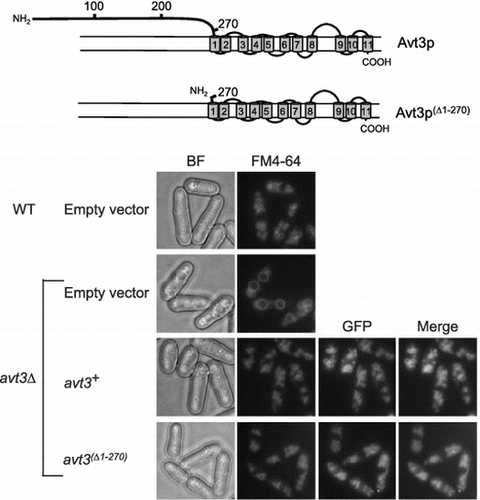
Fig. 1. Topology models of Avt3p and immunoblot analysis of GFP–Avt3p and GFP–Avt3p(Δ1–270) expressed in S. pombe avt3∆ cells.
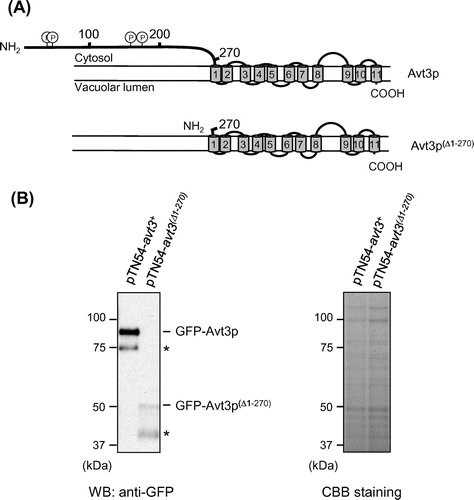
Fig. 2. Effect of deletion of the N-terminal region of Avt3p on its subcellular localization and vacuolar morphology in S. pombe cells.
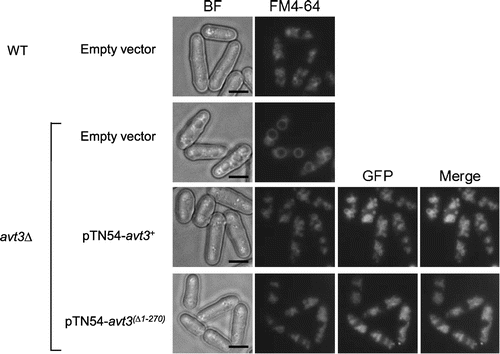
Fig. 3. Effect of deletion of the N-terminal region of Avt3p on vacuolar amino acid composition in S. pombe cells.
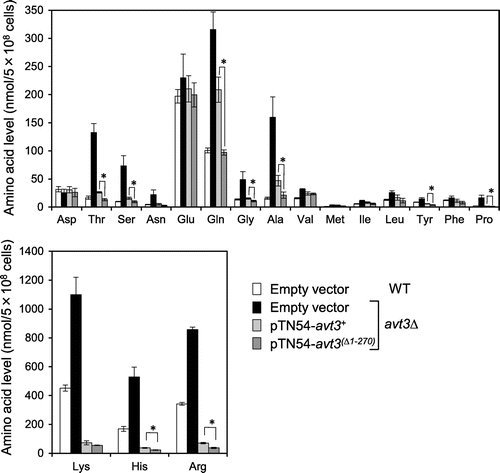
Fig. 4. Effect of deletion of the N-terminal region of Avt3p on spore formation of S. pombe.
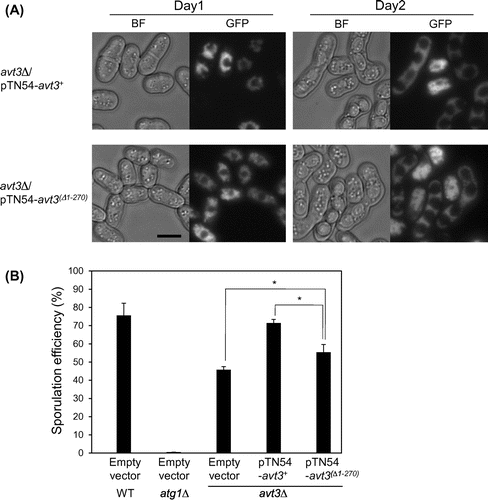
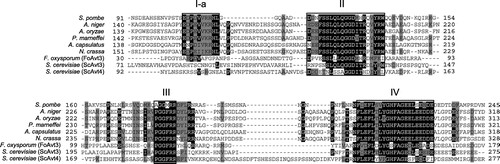
Fig. 5. Conserved sequence elements in the N-terminal hydrophilic regions of fungal Avt3/Avt4 homologs.