Figures & data
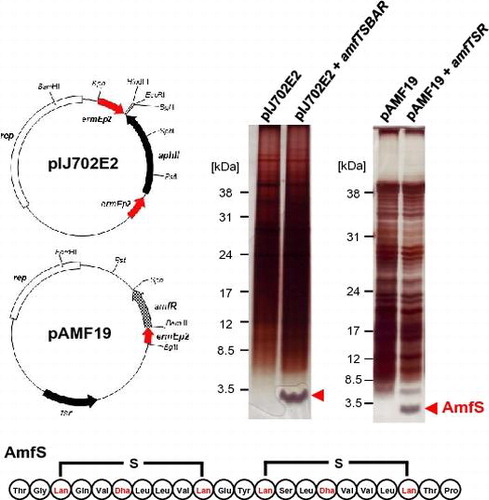
Fig. 1. Schematic representation of the class III lantipeptide biosynthetic gene clusters in Streptomyces and related bacteria and the amino acid sequences of the precursor AmfS.
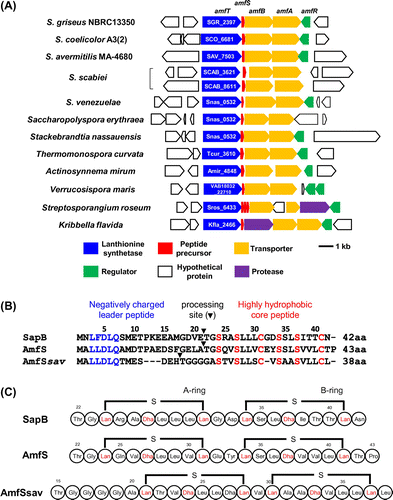
Fig. 2. Phenotype of S. griseus Grd1 mutant.
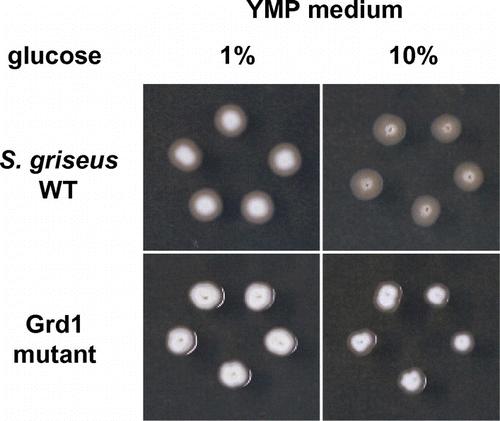
Fig. 3. Physical structures of the expression vectors pIJ702E2 and pAMF19.
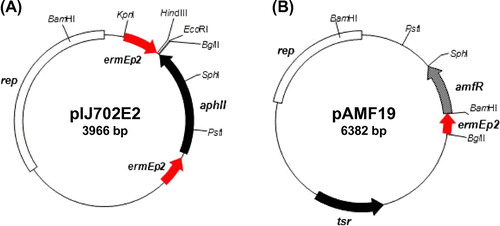
Fig. 4. Schematic representation of AmfS expression vectors and the expression analyses of AmfS in S. griseus by SDS-PAGE.
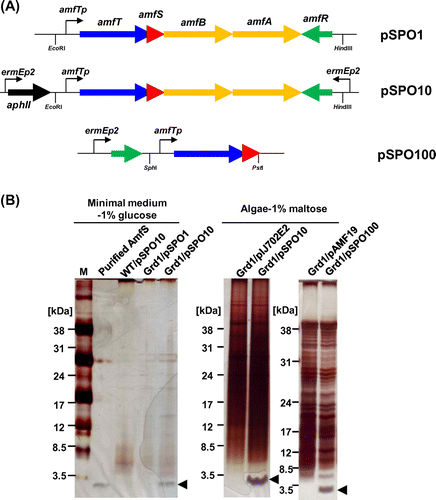
Fig. 5. Morphology of S. avermitilis ΔamfSsav and production of S. avermitilis AmfSsav in the S. griseus Grd1 strain.

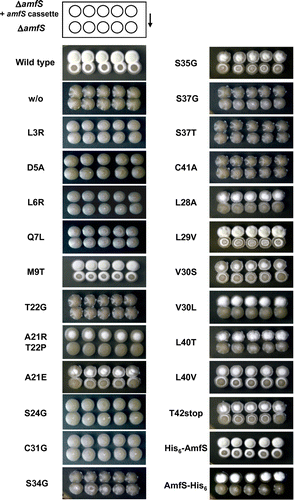
Fig. 6. Genetic and extracellular-complementation tests of S. griseus ΔamfS.