Figures & data
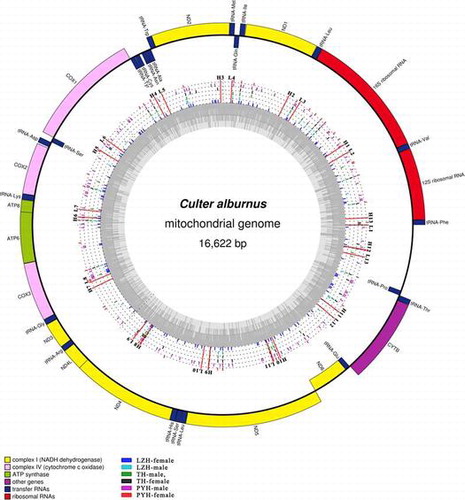
Table 1. PCR primers in the analysis of the C. alburnus mitochondrial genomes.
Fig. 2. Circular representation of the complete mitochondrial genomes of C. alburnus. Circles display (from inside to outside): LZH-female, LZH-male, TH-male, TH-female, PYH-male, and PYH-female. Different color rods and dots represent the variable sites and the same nucleotides, respectively. L1–L13: the location of forward primers. H1–H13: the location of reverse Primer.
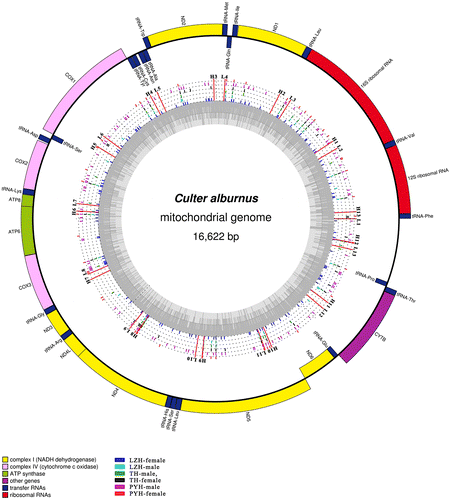
Table 2. Similarity of 16 homolog genes among C. alburnus (retention three significant digits).
Fig. 3. The complete control regions of three geographical groups. Termination associated sequences (TAS) as well as conserved sequence blocks (CSB-D, CSB-E, CSB-F CSB-1, CSB-2, and CSB-3) were underlined. Dots and dashes indicated the same nucleotides and gaps, respectively. Shaded region designated the motif of TGCAT and TACAT. ATGTA were bold. The number on the right side of alignment shows the nucleotide positions.
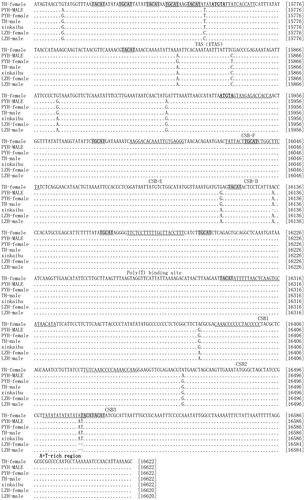
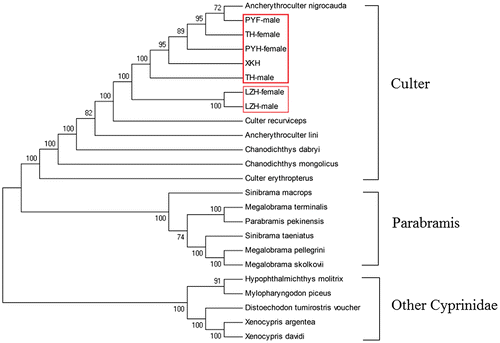
Fig. 4. Phylogenetic analysis based on the complete mitochondrial DNA sequences of various fish. Numbers at tree nodes refers to percent bootstrap values after 1000 replicates. The LZH-female, LZH-male, TH-female, TH-male, PYH-female, and PYH-male were boxed in the phylogenetic tree. Culter alburnus (XKH, KF039719.1), Ancherythroculter lini (KR864861.1), Ancherythroculter nigrocauda (KC513573.1), Chanodichthys dabryi (KC526217.1), Chanodichthys mongolicus (KC701385.1), Culter erythropterus (KJ801524.1), Culter recurviceps (KJ609181.1), Distoechodon tumirostris voucher (DQ026431.1), Hypophthalmichthys molitrix (KJ746944.1), Megalobrama pellegrini (KP262030.1), Megalobrama skolkovii (KJ630486.1), Megalobrama terminalis (AB626850.1), Mylopharyngodon piceus (EU979305.1), Parabramis pekinensis (JX242531.1), Sinibrama macrops (KC122916.1), Sinibrama taeniatus (KM272589.1), Xenocypris argentea (AP009059.1), Xenocypris davidi (KF039718.1).