Figures & data
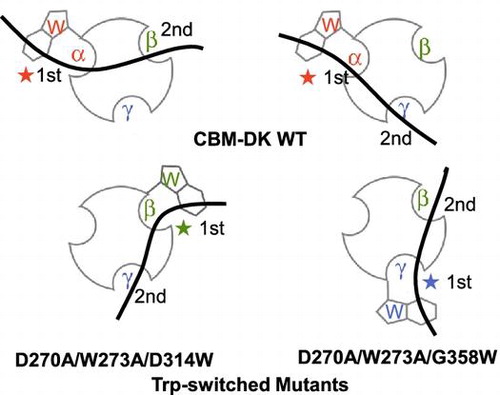
Fig. 1. The amino acid sequence of CBM-DK. The sequence alignments of α-, β-, and γ-repeats were carried out based on the 3D structure model of CBM-DK, reported previouslyCitation9). Single dashes denote identity to α-repeat, and double dashes indicate no amino acid residue at that position.

Fig. 2. Secondary structure and monomeric state analyses. (a) Far-UV CD spectra of CBM-DK wild-type (solid line), D270A/W273A/D314W (dotted line), and D270A/W273A/G358W (broken line). The spectra of other mutants used in this study were also similar. (b) (upper) Sedimentation pattern and best fit result obtained from the c(s) analysis of a solution of CBM-DK wild-type and D270A/W273A/G358W. Residuals between observed data and best-fit result are also indicated. (lower) Distribution of CBM-DK wild-type and D270A/W273A/G358W.
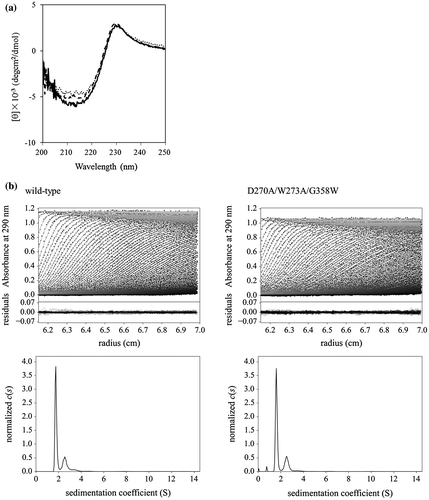
Fig. 3. Typical sensorgrams for the binding of laminarin to D270A/W273A/D314W. (a) D270A/W273A/D314W was immobilized on a CM5 sensorchip and laminarin (0.625–40 μM) was flowed over the chip surface at rate of 20 μl/min for 3 min, followed by 3 min dissociation. (b) Scatchard plots for the laminarin binding. The Ka values determined from the linear fitting to the laminarin in the range of 5–40 μM and 0.625–5 μM, respectively.
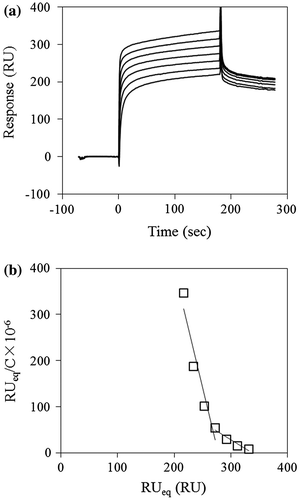
Table 1. Ka values (M−1) for interactions of β-glucans with immobilized CBM-DK and its mutants on the sensor chip.
Fig. 4. Typical ITC data for the binding of laminarin to D270A/W273A/D314W. (a) The 500 μM laminarin solution was injected into the 100 μM D270A/W273A/D314W solution. (b) The data points were obtained by integration of the peaks in (a), corrected for the dilution heat, and plotted against the molar ratio, laminarin to D270A/W273A/D314W. The data were fitted using a nonlinear least-squares method.
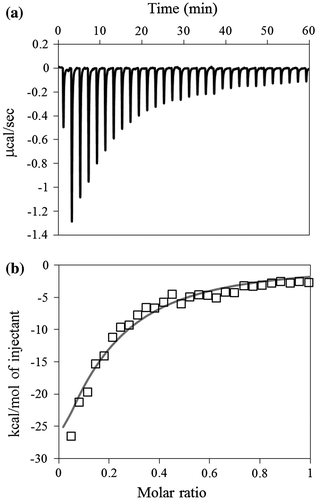
Table 2. Thermodynamic parameters of laminarin binding to CBM-DK and its mutants.
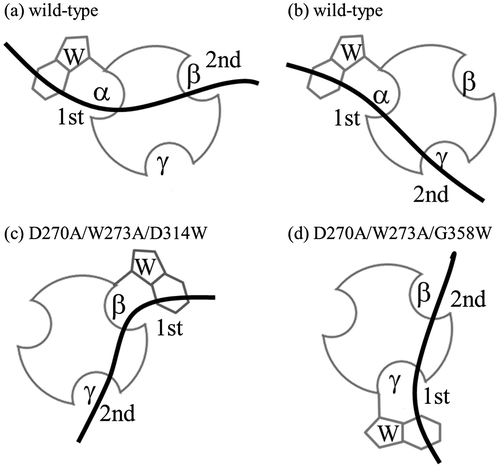
Fig. 5. Schematic representation of binding of CBM-DK wild-type (a, b) and its mutants, D270A/W273A/D314W (c) and D270A/W273A/G358W (d), to laminarin. The curved bold bars and circles with hollows represent laminarin and CBM-DKs, respectively. The Trp-introduced sites and the respective repeats are also indicated. The 1st and 2nd indicate the primary and secondary binding sites for laminarin.