Figures & data
Fig. 1. The amounts of n-hexanal (A) and (Z)-3-hexenal (B) formed from intact or mechanically wounded thalli, antheridiophores, and archegoniophores.
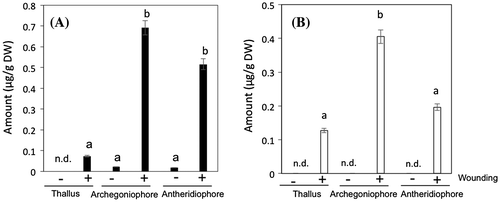
Fig. 2. Phylogenetic analysis of MpLOXs with the LOXs from C. reinhardtii, K. flaccidum, Marchantia polymorpha, P. patens, S. moellendorffii, A. thaliana, Z. mays, P. trichocarpa, and P. abies.
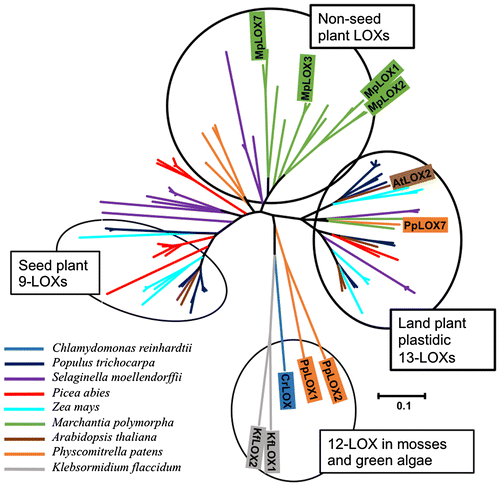
Fig. 3. Amino acid sequence of MpLOX7.
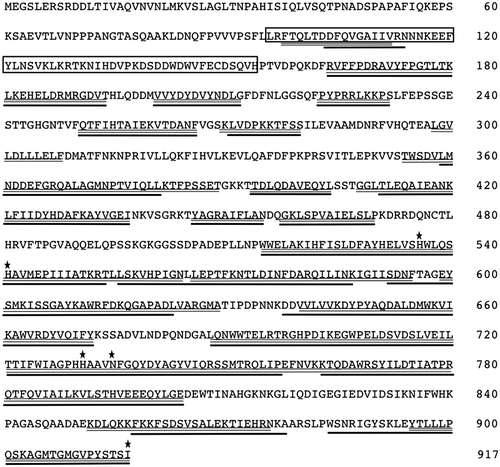
Table 1. Kinetic parameters and main products of recombinant MpLOX7 with fatty acid substrates.
Fig. 4. Formation of n-hexanal from arachidonic acid (ARA) (A) and (Z)-3-hexenal from α-linolenic acid (LNA) (B) during catalysis of recombinant MpLOX7.
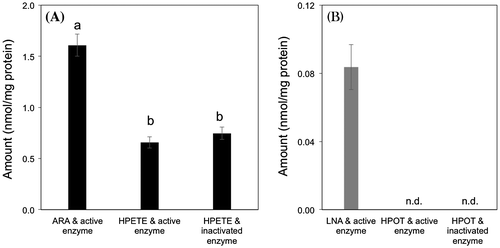
Fig. 5. Expression level of MpLOX7 in thalli, archegoniophores before mechanical wounding and after mechanical wounding, and antheridiophore before mechanical wounding and after mechanical wounding.
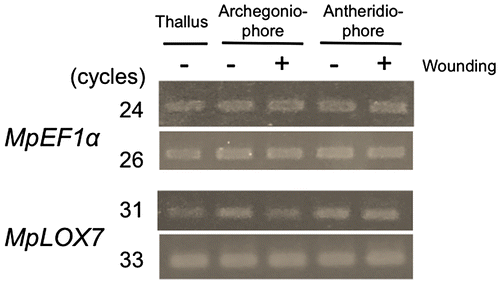
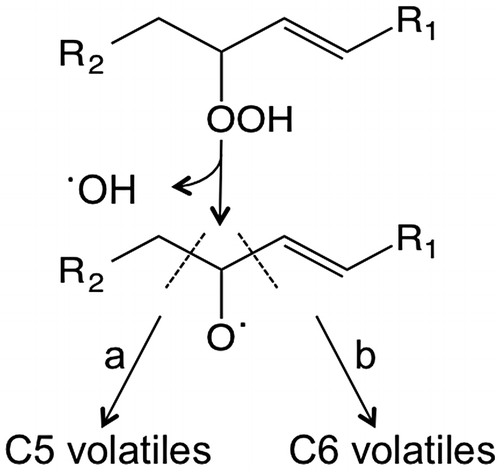
Fig. 6. Proposed routes of β-scission of fatty acid hydroperoxides formed by lipoxygenases.