Figures & data
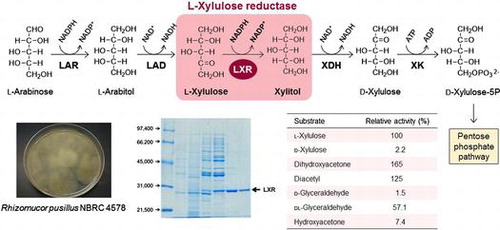
Fig. 1. l-Arabinose and d-xylose catabolism pathways in fungi and yeasts. LAR; l-arabinose reductase, LAD; l-arabinitol dehydrogenase, LXR; l-xylulose reductase, XR; d-xylose reductase, XDH; xylitol dehydrogenase.
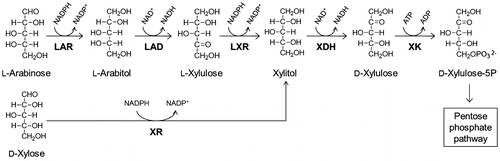
Table 1. Purification steps of native RpLXR.
Fig. 2. SDS-PAGE of RpLXR during purification. Lanes. 1, Molecular-weight standards; 2, Cell-free extract; 3, Ammonium sulfate fractionation (50–90%); 4, Butyl-Toyopearl column chromatography; 5, Gigapite column chromatography; 6, MonoQ HR 5/5 column chromatography; 7, Superdex 200 HR 10/30 column chromatography.
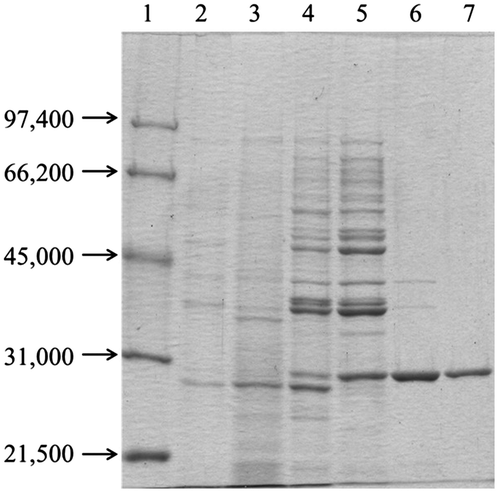
Table 2. Substrate specificity of the native RpLXR.
Table 3. Comparison of l-xylulose reductase from several fungi.
Fig. 3. Phylogenetic tree of l-xylulose reductase from R. pusillus NBRC 4578 and its homologous proteins. The phylogenetic tree was constructed using ClustalW and MEGA7. The arrow showed RpLXR sequence determined in this study.
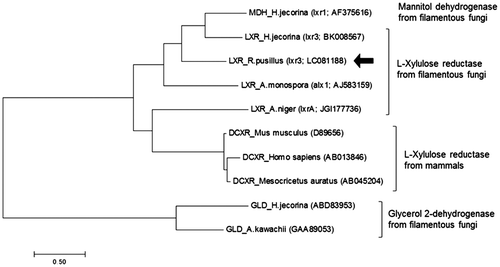
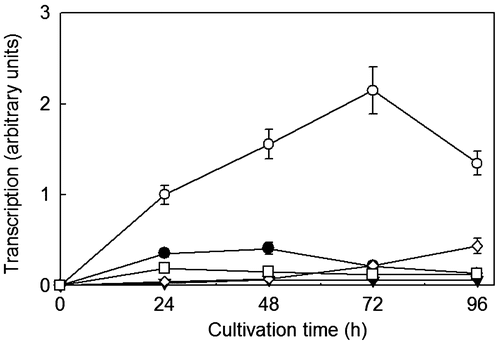
Fig. 4. Expression of the Rplxr3 gene in R. pusillus NBRC 4578 cultured on d-glucose (▼), l-xylose (●), l-arabinose (○), d-mannose (□), and glycerol (◊). The expression level of mycelia cultivated for 24 h on L-arabinose set as 1.0.