Figures & data
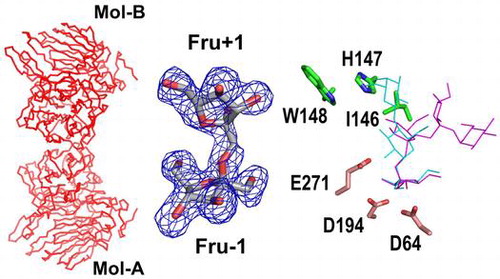
Table 1. Data collection and refinement statistics.
Figure 1. Enzymatic properties of AkFFase.
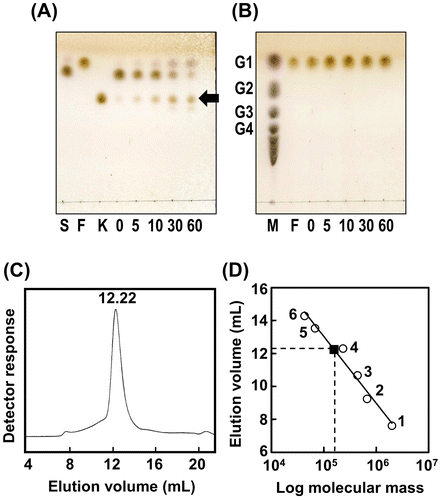
Figure 2. Overall structure of AkFFase.
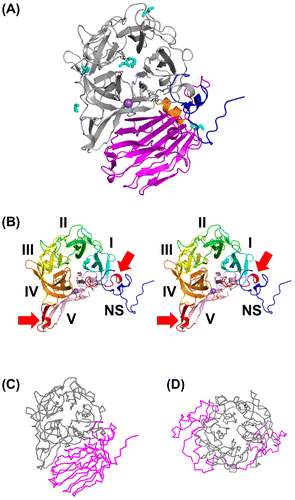
Table 2. Summary of structural similarity search using the DALI server.
Figure 3. Comparison of the Cα backbones of AkFFase and related GH32 enzymes.
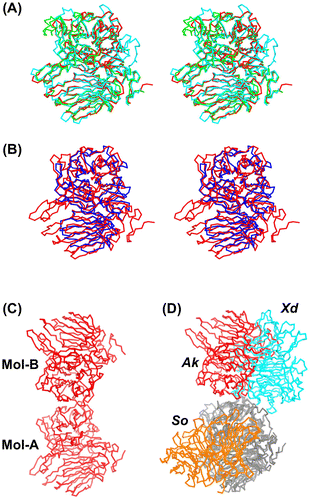
Figure 4. Structure of the AkFFase-Fru complex.
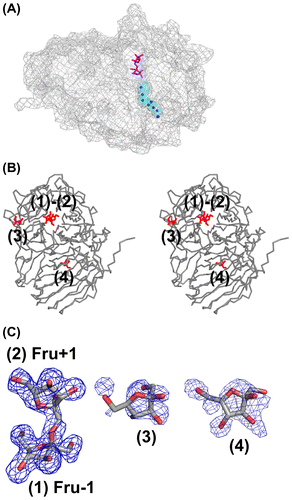
Figure 5. Schematic drawing of the amino acid residues interacting with Fru −1 and Fru +1.
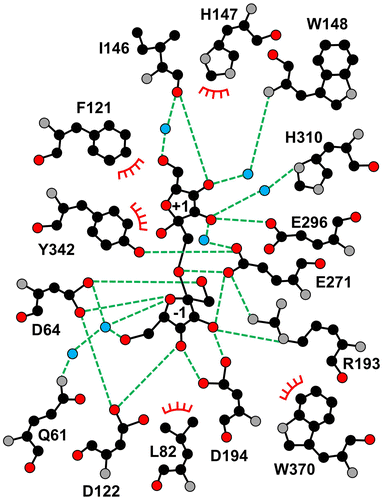
Table 3. Amino acid residues in the catalytic cleft of AkFFase and the corresponding residues in other clan GH-J enzymes.
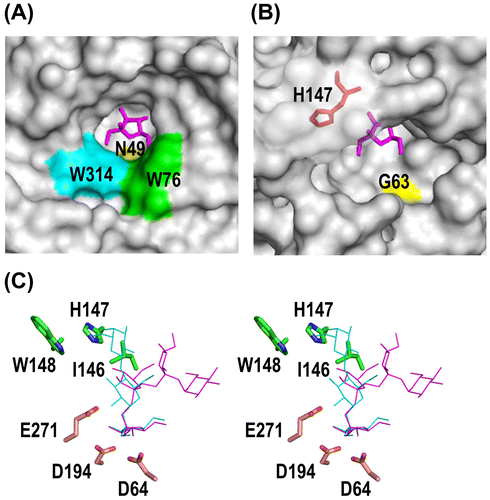
Figure 6. Structure of the catalytic cleft of AkFFase.