Figures & data
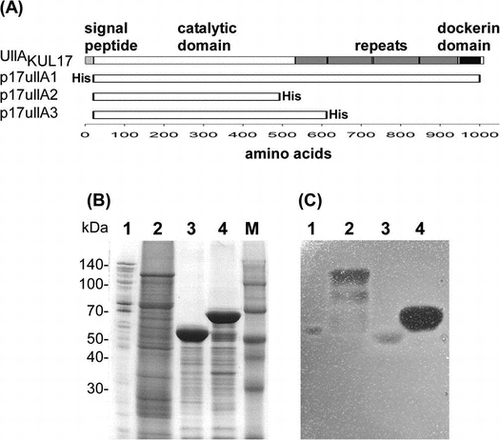
Table 1. Primers and plasmids used in this study.
Fig. 1. SDS-PAGE analysis of proteins separated by phenyl Sepharose chromatography.
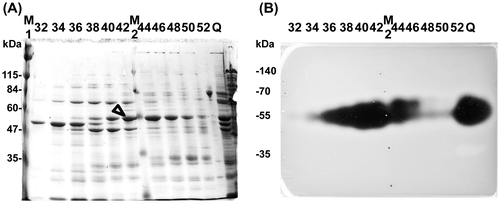
Fig. 2. Identification of ulvan-degrading enzyme.
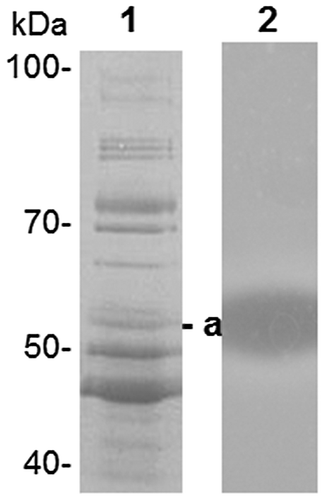
Fig. 3. Domain structure and purification of UllA.
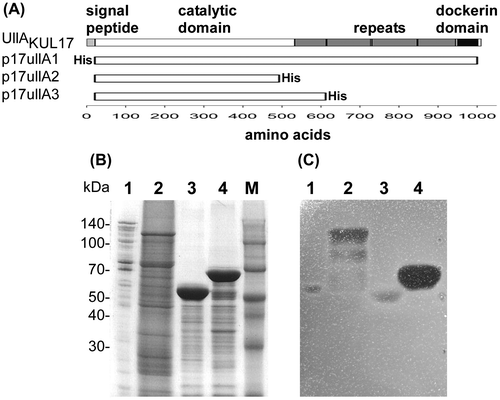
Fig. 4. Enzyme assay of UllA1.
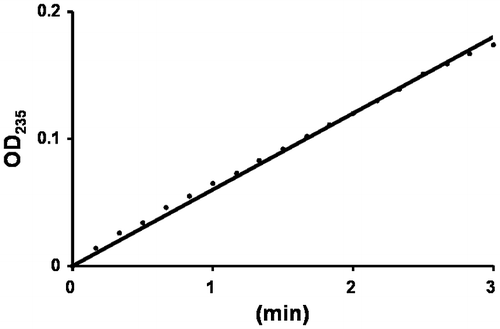
Fig. 5. Analysis of reaction products of ulvan by UllA3.
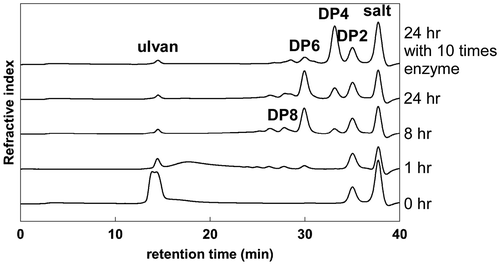
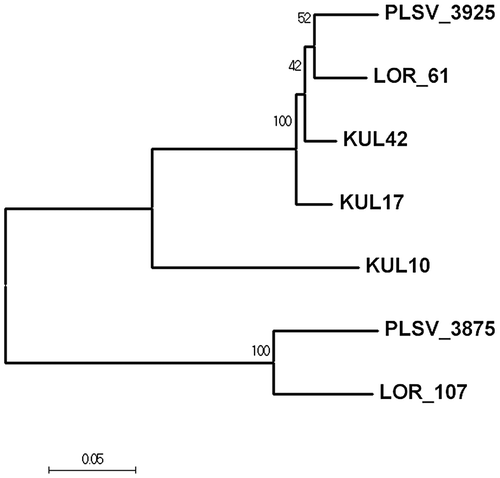
Fig. 6. Evolutionary relationship of long-type ulvan lyase. The evolutionary history was inferred using the Neighbor-Joining method.Citation30)