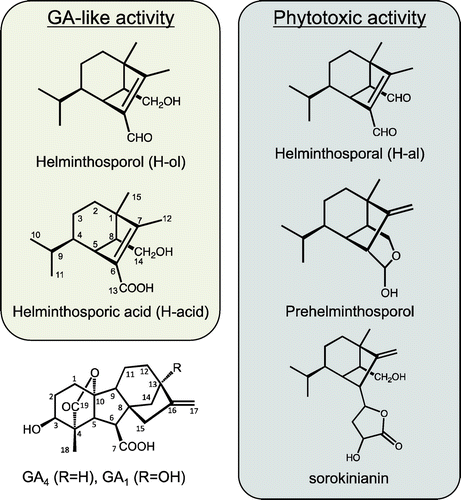
Figures & data
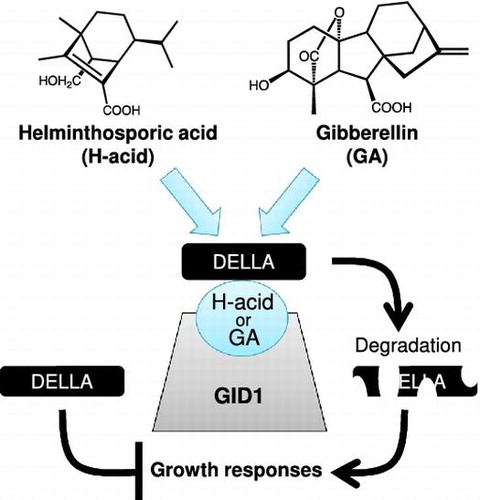
Figure 2. GA-like activity of H-acid in rice and Arabidopsis.
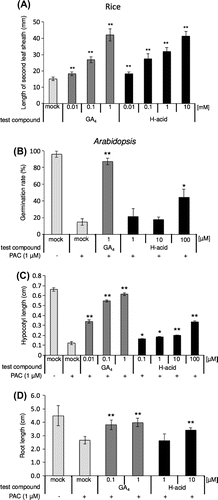
Figure 3. H-acid regulates the expression of GA-related genes in the same manner as GA4.
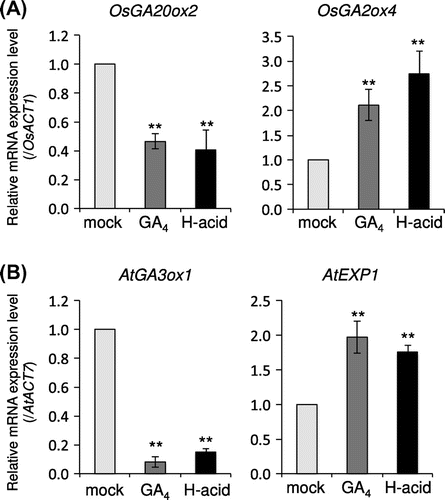
Figure 4. H-acid promoted the degradation of Arabidopsis DELLA protein.
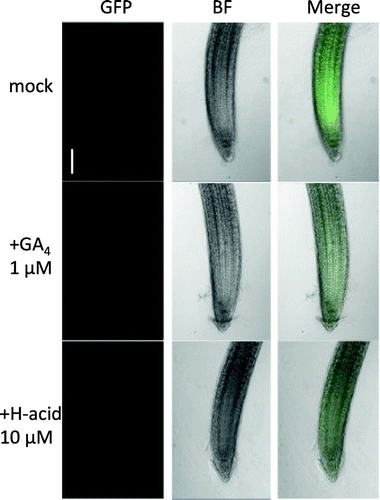
Figure 5. H-acid promotes the formation of GID1-DELLA complex.
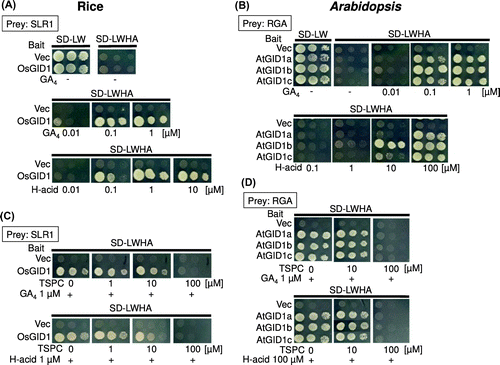
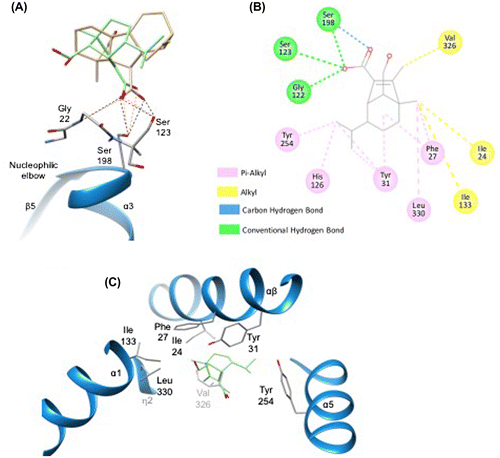
Figure 6. Docking simulation analysis of interaction between H-acid and OsGID1.