Figures & data
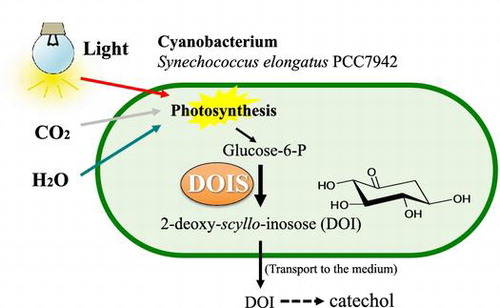
Table 1. Oligonucleotide primers used in this study.
Figure 1. IPTG-dependent expression of the btrC gene at the chromosomal neutral site. (A) Schematic diagram of the btrC-expressing gene construct in the chromosomal neutral site of S. elongatus (NS-btrC). The btrC gene under the control of the trc promoter was introduced into the neutral site of the S. elongatus chromosome. btrC, the gene coding catalytic subunit of 2-deoxy-scyllo-inosose synthase (DOIS); Spr, the spectinomycin resistance marker gene; lacIq, the repressor gene for the trc promoter. ((B), (C)) Expression of the BtrC protein without (left) or with IPTG (right). The cells were grown to the logarithmic phase at 30 °C under standard lighting (40 μmol photon m−2 s−1), and btrC was induced by IPTG (final concentration 1 mM). Crude extracts were prepared 24 h after the addition of IPTG, and samples (10 μg protein) were analyzed by SDS-PAGE (B) and western blotting using an anti-BtrC antibody (C). The arrows indicate the BtrC protein signals.
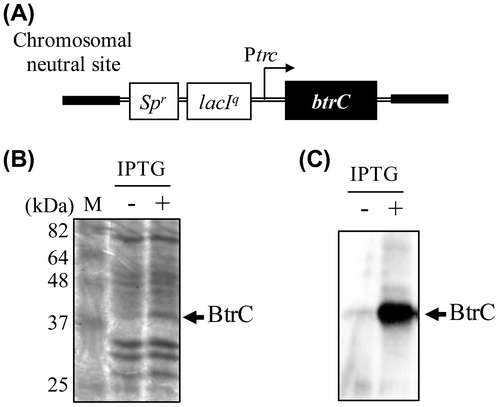
Figure 2. Intensive and constitutive expression of the BtrC protein in the plasmid. (A) Scheme of the btrC-expressing construct in the pEX1 (pEX1-btrC plasmid). The gene construct, containing the btrC gene under the trc promoter, was cloned into the XbaI/XhoI site in the plasmid vector pUC303, and introduced to the S. elongatus. cat, the chloramphenicol resistance marker gene. BtrC was constitutively expressed because the pEX1-btrC plasmid does not contain the lacIq gene. (B) Expression of the BtrC protein. The cells were grown to the logarithmic phase at 30 °C under standard lighting with bubbling. Crude extracts were prepared 24 h after inoculation, and samples (10 μg protein) were analyzed by SDS-PAGE. (C) Growth of S. elongatus expressing BtrC. The growth curves of S. elongatus PCC 7942 containing pUC303 (closed squares represent the empty vector) and the pEX1-btrC plasmid (open diamonds) are shown.
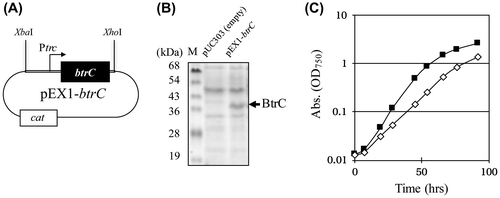
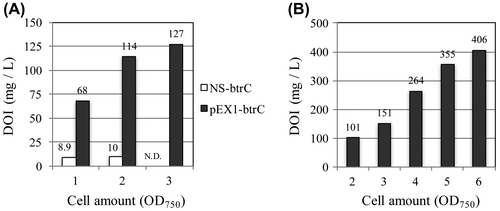
Figure 3. Production of 2-deoxy-scyllo-inosose (DOI) in BtrC-expressing strains. (A) S. elongatus strains containing NS-btrC and pEX1-btrC were grown at 30 °C under standard lighting (40 μmol photon m−2 s−1) while bubbling air through the medium. When the optical density (OD750), which indicates the number of cells in the culture, reached the indicated value, the supernatant of the culture was harvested, and the amount of DOI was determined by HPLC. (B) As the S. elongatus PCC 7942 strain containing the pEX1-btrC plasmid was cultured, the light intensity was increased in stepwise increments (75–400 μmol photon m−2 s−1, see Materials and Methods). At each step, the supernatant of the culture was harvested, and the amount of DOI was determined.