Figures & data
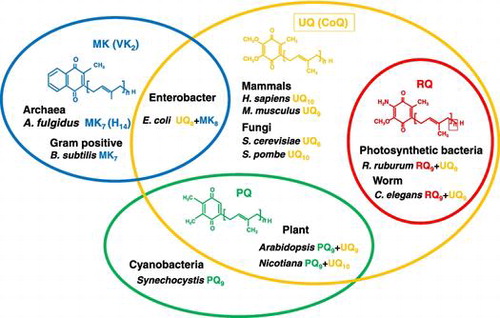
Figure 1. Distribution of four major prenylquinones in different organisms. Ubiquinone (UQ) is present in almost all living organisms from bacteria to higher eukaryotes. Menaquinone (MK) is distributed in bacteria and archaea, and therefore considered the oldest type of prenylquinone, first synthesized in primitive living organisms. Plastoquinone (PQ) occurs in cyanobacteria, and was presumably subsequently transferred to plants. Rhodoquinone (RQ) is the most recently evolved quinone, and is synthesized from UQ.
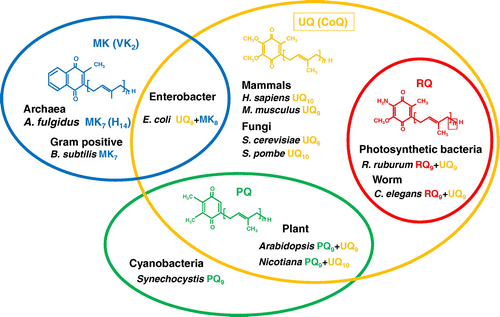
Figure 2. The mevalonate (MVA) pathway in Saccharomyces cerevisiae. The MVA pathway consists of seven enzyme-catalyzed reactions. The first step is the formation of acetoacetyl-CoA from two acetyl-CoA molecules by Erg10 (acetyl-CoA acetyl transferase). Subsequently, Erg13 (HMG-CoA synthase), Hmg1/Hmg2 (HMG-CoA reductase), Erg12 (mevalonate kinase), Erg8 (phosphomevalonate kinase), and Erg19 (diphosphomevalonate decarboxylase) lead to the production of isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP). Idi1 isomerizes between IPP and DMAPP.
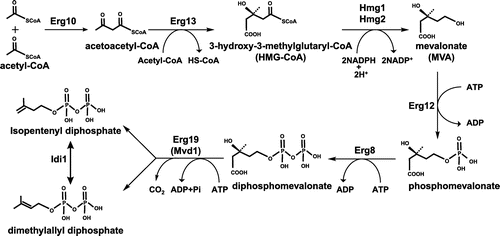
Figure 3. The 2C-methyl-D-erythritol-4-phosphate (MEP) pathway in Escherichia coli. The MEP pathway consists of eight enzyme-catalyzed reactions. The first step is the condensation of pyruvate and glyceraldehyde-3-phosphate to form 1-deoxy-D-xylulose-5-phosphate by Dxs (DXP synthase). Subsequent steps catalyzed by IspC (DXP reductoisomerase), IspD (MEP cytidyltransferase), IspE (CDP-ME kinase), IspF (MECDP synthase), IspG (4-hydroxy-3-methylbut-2-enyl diphosphate synthase), and IspH (4-hydroxy-3-methylbut-2-enyl diphosphate reductase) lead to the production of IPP and DMAPP.
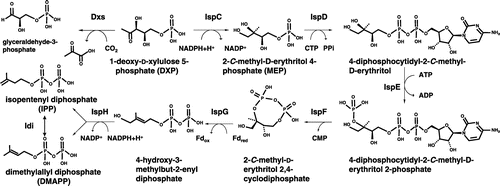
Figure 4. Biosynthetic pathway of the isoprenoid tail of prenylquinones. Polyprenyl diphosphate synthase synthesizes trans-polyprenyl diphosphate of a certain length. S. cerevisiae Coq1 (hexaprenyl diphosphate synthase) forms products from six isoprene units, E. coli IspB (octaprenyl diphosphate synthase) synthesizes products with eight isoprene units, Arabidopsis SPS1, SPS2, and SPS3 (solanesyl diphosphate synthase) generate products with nine isoprene units, and human and Schizosaccharomyces pombe decaprenyl diphosphate synthase (DPS; a heteromer of PDSS1 and PDSS2 or Dps1 and Dlp1, respectively) catalyzes the formation of products with ten isoprene units. S. cerevisiae Coq2 (PHB-hexaprenyl diphosphate transferase), E. coli UbiA (PHB-octaprenyl diphosphate transferase), and human COQ2 (PHB-decaprenyl diphosphate transferase) or S. pombe Ppt1 (Coq2; PHB-decaprenyl diphosphate transferase) condense p-hydroxybenzoate (PHB) with trans-polyprenyl diphosphate to form UQ6, UQ8, and UQ10, respectively. MenA prenylates DHNA, and homogentisate solanesyl transferase (HST) prenylates homogentisate (HGA). DPP, decaprenyl diphosphate; HexPP, hexaprenyl diphosphate; NPP, nonaprenyl diphosphate; OPP, octaprenyl diphosphate.
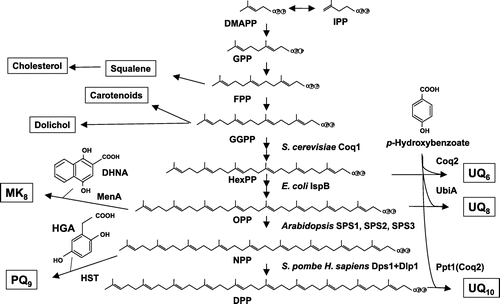
Figure 5. Overview of the proposed UQ biosynthetic pathway. The UQ biosynthetic pathways of E. coli and S. cerevisiae are shown. In E. coli, PHB is first condensed with trans-polyprenyl diphosphate, and the ring structure is then modified. Decarboxylation catalyzed by UbiD (3-octaprenyl-4-hydroxybenzoate decarboxylase) and UbiX (flavin prenyl transferase) follows. The ring is further hydroxylated by UbiI (2-octaprenylphenol hydroxylase), O-methylated by UbiG (2-octaprenyl-6-hydroxy phenol methylase), hydroxylated by UbiH (2-octaprenyl-6-methoxyphenol hydroxylase), C-methylated by UbiE (2-octaprenyl-6-methoxy-1,4-benzoquinone methylase), hydroxylated by UbiF (2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinone oxygenase), and O-methylated by UbiG (3-demethylubiquinone 3-methyltransferase). In S. cerevisiae, para-amino benzoic acid (pABA) and PHB are used for UQ synthesis. The first ring is modified via hydroxylation by Coq6 (PHB-2-hexaprenyl hydroxylase), followed by O-methylation by Coq3 (2-hexaprenyl-6-hydroxy phenol methyltransferase). After decarboxylation and hydroxylation steps, the ring is further modified via C-methylation by Coq5 (2-hexaprenyl-6-methoxy-1,4-benzoquinone methyltransferase), a final hydroxylation by Coq7 (2-hexaprenyl-3-methyl-6-methoxy-1,4-benzoquinone oxygenase), and O-methylation by Coq3. H. sapiens contains similar enzymes with S. cerevisiae except PDSS1 and PDSS2 [Citation70].
![Figure 5. Overview of the proposed UQ biosynthetic pathway. The UQ biosynthetic pathways of E. coli and S. cerevisiae are shown. In E. coli, PHB is first condensed with trans-polyprenyl diphosphate, and the ring structure is then modified. Decarboxylation catalyzed by UbiD (3-octaprenyl-4-hydroxybenzoate decarboxylase) and UbiX (flavin prenyl transferase) follows. The ring is further hydroxylated by UbiI (2-octaprenylphenol hydroxylase), O-methylated by UbiG (2-octaprenyl-6-hydroxy phenol methylase), hydroxylated by UbiH (2-octaprenyl-6-methoxyphenol hydroxylase), C-methylated by UbiE (2-octaprenyl-6-methoxy-1,4-benzoquinone methylase), hydroxylated by UbiF (2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinone oxygenase), and O-methylated by UbiG (3-demethylubiquinone 3-methyltransferase). In S. cerevisiae, para-amino benzoic acid (pABA) and PHB are used for UQ synthesis. The first ring is modified via hydroxylation by Coq6 (PHB-2-hexaprenyl hydroxylase), followed by O-methylation by Coq3 (2-hexaprenyl-6-hydroxy phenol methyltransferase). After decarboxylation and hydroxylation steps, the ring is further modified via C-methylation by Coq5 (2-hexaprenyl-6-methoxy-1,4-benzoquinone methyltransferase), a final hydroxylation by Coq7 (2-hexaprenyl-3-methyl-6-methoxy-1,4-benzoquinone oxygenase), and O-methylation by Coq3. H. sapiens contains similar enzymes with S. cerevisiae except PDSS1 and PDSS2 [Citation70].](/cms/asset/ad26797c-6ee4-4674-87eb-bf2e0e39bcf2/tbbb_a_1433020_f0005_oc.gif)
Figure 6. Menaquinone biosynthesis in E. coli. The biosynthesis of menaquinone in E. coli starts from the conversion of chorismate to isochorismate by MenF. Subsequently, MK is synthesized by MenD (succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexadiene-1-carboxylate synthase), MenH (2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase), MenC (O-succinylbenzoate synthase), MenE (O-succinylbenzoic acid-CoA ligase), MenB (naphthoate synthase), MenI (DHNA-CoA thioesterase), MenA (1,4-dihydroxy-2-naphthoate octaprenyltransferase), and MenG (UbiE). *MenJ functions as a reductase of the side chain in Mycobacterium tuberculosis.
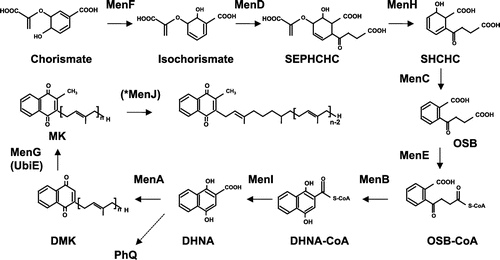
Figure 7. Novel pathway of menaquinone synthesis via futalosine. A novel menaquinone biosynthesis pathway was originally discovered in Streptomyces coelicolor, in which 1,4-dihydroxy-6-naphthoate is synthesized by MqnA (chorismate dehydratase), MqnD (1,4-dihydroxy-6-naphthoate synthase), MqnE (aminofutalosine synthase), AFL deaminase, MqnB (futalosine hydrolase), and MqnC (dehypoxanthine futalosine cyclase). MK synthesis from 1,4-dihydroxy-6-naphthoate is still not clearly understood, but a prenylation step catalyzed by MqnP has been proposed.
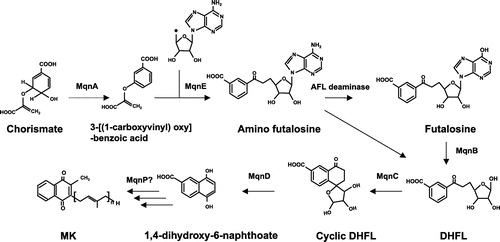
Figure 8. Crystal structures of menaquinone biosynthetic enzymes. (A) MenF from E. coli (PDB ID: 2EUA). (B) MenC from E. coli (PDB ID: 1FHU). (C) MenB from E. coli (PDB ID: 3T89). (D) MenD from E. coli (PDB ID: 3HWX). (F) MenE from E. coli (PDB ID: 5C5H). (G) MenH from E. coli (PDB ID: 4GDM). (H) MenI from E. coli (PDB ID: 4K4B). (I) MqnA from Deinococcus radiodurans (PDB ID: 216E). (J) MqnD from Thermus thermophilus HBB (PDB ID: 3A3U). (K) MqnB from Helicobacter pylori (PDB ID: 4BMX).
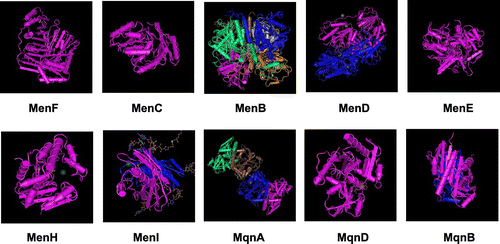
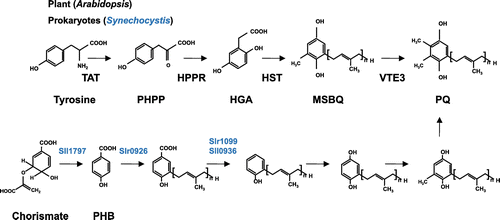
Figure 9. The biosynthesis of plastoquinone. In plants, tyrosine is converted to p-hydroxyphenylpyruvate (PHPP) by TAT (tyrosine amino transferase), and the product is oxidized to HGA by HPPD (p-hydroxyphenylpyruvate dioxygenase). HGA is prenylated by HST (homogentisate prenyltransferase), and methylated by VTE3 (MSBQ methyltransferase) to yield PQ. In Synechocystis, PHB is used as a quinone backbone, and after prenylation of PHB by Slr0926, decarboxylation, oxidation, and methylation take place.