Figures & data
Table 1. Oligonucleotide primers used for the cloning of mSULT2A8 and the construction of mutant mSULT2A8.
Figure 2. Phylogenic tree analysis of mouse SULTs on the basis of their amino acid sequences. Notes: The dendrogram shows the degree of amino acid sequence homology among mouse SULTs. Protein sequences were extracted from the GeneBank database and accession number of individual SULTs are as follows: mSULT1A1(NP_598431), mSULT1B1 (NP_063931), mSULT1C1 (NP_061221), mSULT1C2 (NP_081211), mSULT1D1 (NP_058051), mSULT1E1 (NP_075624), mSULT2A1 (NP_001104766), mSULT2A2 (AAA40145), mSULT2A3 (NP_001095056), mSULT2A4(NP_001095004), mSULT2A5 (NP_001171909), mSULT2A6 (NP_001074794), mSULT2A7 (modified from LOC194586), mSULT2A8 (NP_780459), mSULT2B1a1(XP_006541060), mSULT2B1a2(XP_006541061), mSULT2B1b (NP_059493), mSULT3A1(NP_065590), mSULT3A2 (NP_001094922), mSULT4A1 (NP_038901), mSULT5A1 (NP_065589), mSULT6B1 (NP_001157097). Construction of dendrogram was performed based on the methods as described in “Materials and Methods”.
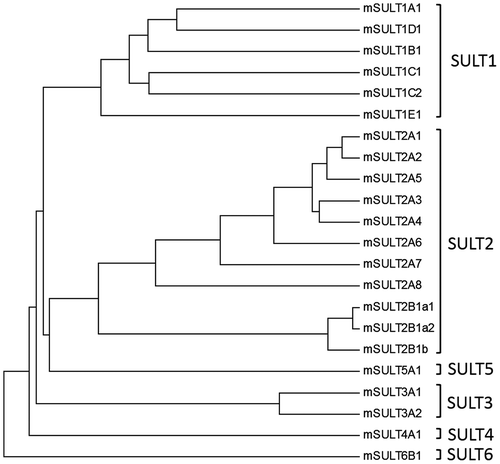
Figure 3. Amino acid sequence alignment analysis of human and mouse SULT2A subfamily members. Notes: Identical residues conserved among at least two of the nine SULT enzymes are drawn in Black, and similar residues are in gray. 5′-PSB loop and 3′-PB motif located, respectively, in the N-terminal and middle regions are underlined. All SULTs, except mSULT2A8, contain His99 residue in their amino acid sequences as indicated by solid arrow. His48 residue of mouse SULT2A8, as indicated by boxed arrow, is also unique as compared with other SULTs.
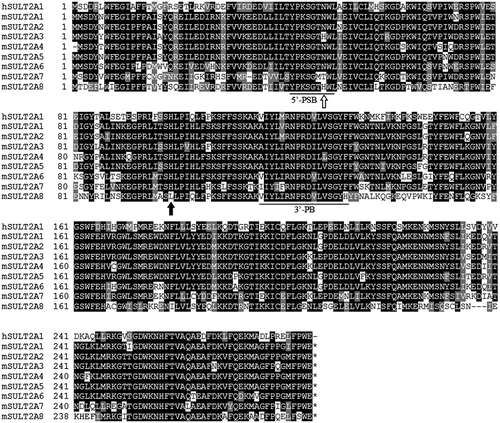
Table 2. Specific activity of mSULT2A8 with bile acids and steroids a.
Table 3. Kinetic parameters of the sulfation of cholic acids by mSULT2A8 a.
Table 4. Specific activity of mutant mSULT2A8 with cholic acids and steroidsa.
Table 5. Kinetic parameters of the sulfation of cholic acid by wild-type and mutant mSULT2A8a.