Figures & data
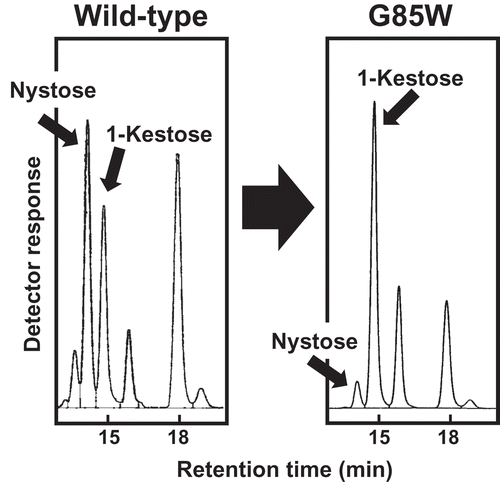
Table 1. Plasmids and oligonucleotide primers used.
Figure 1. HPLC analysis of the products from the incubation of sucrose with E. coli cell suspensions expressing (a) PgsA-AkFFase, (b) PgsA-AoFFase, (c) PgsA-AtFFase, and (d) PgsA-AkFFase G85W.
FN, fructosylnystose; N, nystose; K, 1-kestose; S, sucrose; G, glucose; F, fructose.
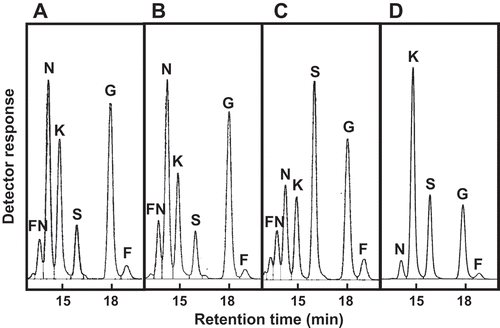
Figure 2. Amount of product from the incubation of sucrose with E. coli cell suspensions expressing PgsA-AkFFase, PgsA-AoFFase, PgsA-AtFFase, and PgsA-AkFFase G85W.
Black bar, 1-kestose; Gray bar, nystose. Error bars represent SEM (standard error of the mean). Each bar of PgsA-AkFFase and PgsA-AkFFase G85W corresponds to the average of four experiments, and each bar of PgsA-AoFFase and PgsA-AtFFase corresponds to the average of two experiments.
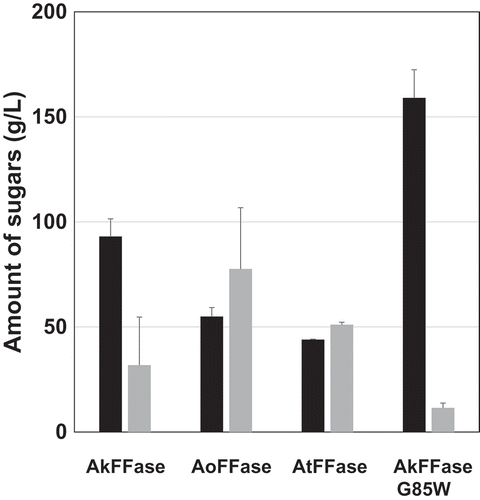
Figure 3. Stereo view of the catalytic cleft of AkFFase. The model of nystose (green) is obtained from PDB entry 3LEM, and placed on the structure of AkFFase (PDB ID, 5XH8).
Subsite numbers are indicated from −1 to +3.
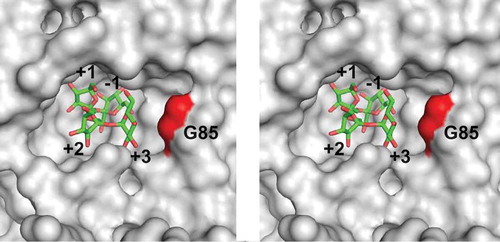
Figure 4. Time courses of the amounts of sucrose, 1-kestose, and nystose in the reaction using E. coli cell expressing PgsA-AkFFase wild-type enzyme (dotted lines) and PgsA-AkFFase G85W mutant (solid lines).
Symbols: white square, wild-type and sucrose; black square, G85W and sucrose; white circle; wild-type and 1-kestose; black circle, G85W and 1-kestose; white triangle, wild-type and nystose; black triangle, G85W and nystose.
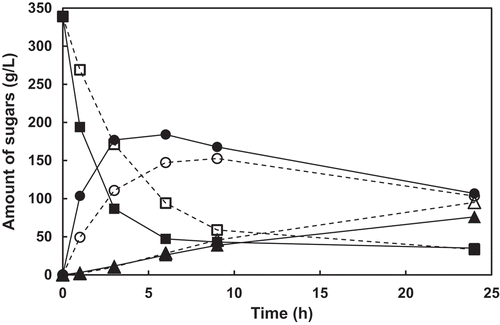
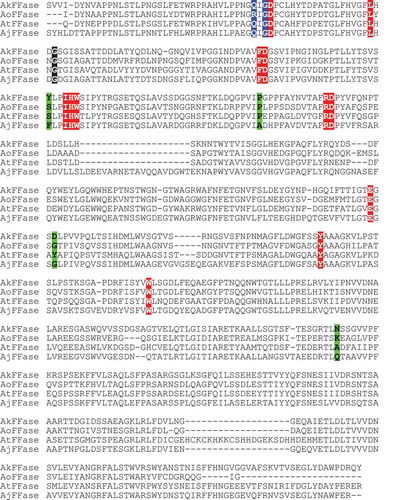
Figure 5. Comparison of amino acid sequences of AkFFase, AoFFase, AtFFase, and A. japonicus FFase (AjFFase).
The sequences were aligned using ClustalOmega server. Colors: red, residues in subsites −1 and +1; black, Gly85 in AkFFase and the corresponding residues; blue, residues located inside the catalytic cleft other than those shown in red or black; green, residues that located distantly from the catalytic site but reportedly influence transfructosylation.