Figures & data
Figure 1. Cyclopiazonic acids produced by fungus Aspergillas oryzae HMP-F28 induced extracellular alkalinization (A) and H2O2 production (B) in tobacco cell suspensions.
SA: salicylic acid; RFI: relative fluorescence intensity. SA and harpin were used as positive controls. Data are means and standard deviations calculated from three independent experiments.
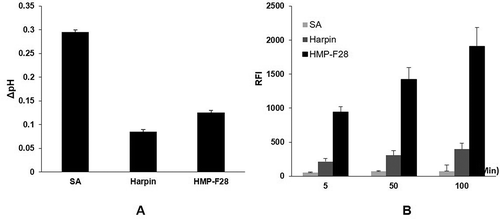
Figure 2. Structure, 2D NMR correlations and structural model analysis of the cyclopiazonic acids (1–4) isolated from Aspergillas oryzae HMP-F28.
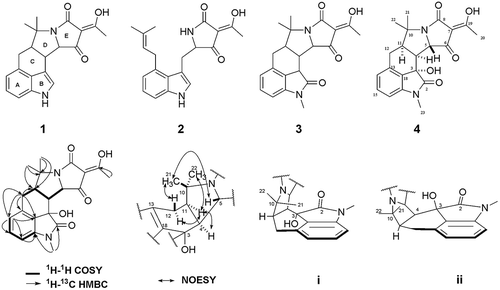
Table 1. NMR data assignments for 3-hydroxysperadine A (4)a.
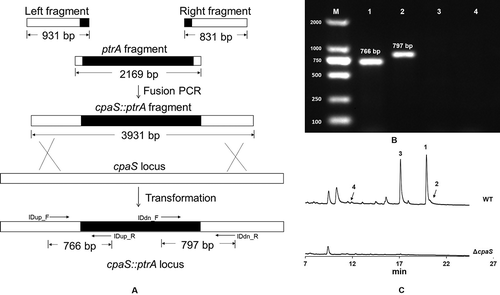
Figure 3. Disruption of the cpaS gene in Aspergillas oryzae HMP-F28.
A: Homologous recombination scheme for inactivation of cpaS gene; B: Amplification of the fragments containing the two boundaries of the inserted ptrA gene and the two arms. The mutant ΔcpaS gave upstream and downstream amplicons with the expected sizes at 766 bp (lane 1) and 797 bp (lane 2), respectively. The wild type didn’t show any specific amplified PCR products; C: Metabolic profiles (UV 285 nm) of the wild-type A. oryzae HMP-F28 (upper panel) and the ΔcpaS mutant (A. oryzae HMP-F28M01, lower panel) harboring a disrupted pks-nrps gene essential for cyclopiazonic acid biosynthesis. Mutant ΔcpaS no longer produced cyclopiazonic acids 1 ~ 4.