Figures & data
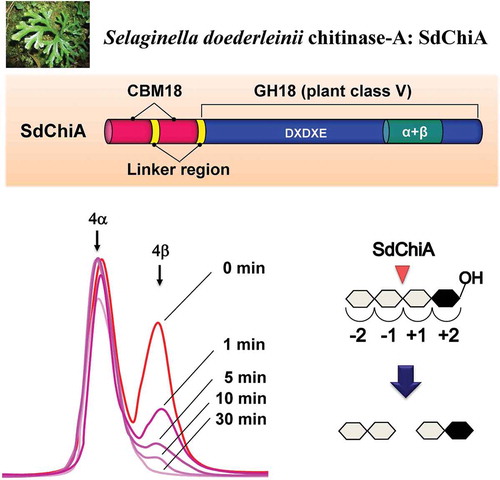
Figure 1. Molecular mass of SdChiA from S. doederleinii.
SDS-PAGE was performed using the method of Laemmli. Lane 1, marker proteins; lane 2, purified SdChiA from S. doederleinii. The gel was stained with Coomassie brilliant blue R250.
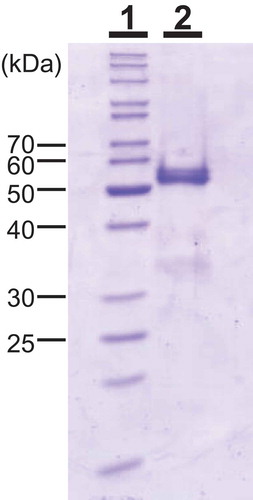
Table 1. Primers for PCR and RACE.
Figure 2. Effect of pH and temperature on the chitinase activity of SdChi-A.
The effect of pH (a) and temperature (b) on activity was examined after incubation at 37°C for 15 min. Methods are described in Materials and Methods.
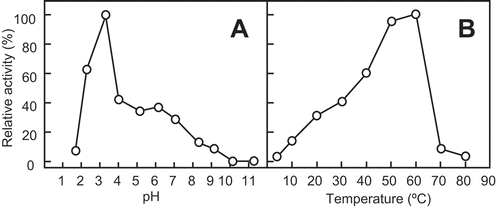
Figure 3. HPLC analysis of the hydrolysis products of (GlcNAc)n by SdChiA.
S, Standards (GlcNAc)1–6. (a) hydrolysis products of (GlcNAc)6; (b) hydrolysis products of (GlcNAc)5; (c) hydrolysis products of (GlcNAc)4. I to VI, α and β indicates (GlcNAc)1–6, α- and β- anomer, respectively. Standard (GlcNAc)1–6 and the reaction mixtures were analyzed by HPLC on a TSKgel amide-80 column as described in Materials and Methods. (d) Schematic representation of the substrate binding mode in the subsites. (GlcNAc) are shown as hexagon. The reducing-end sugars are filled in black or gray. The arrows indicate the cleavage site.
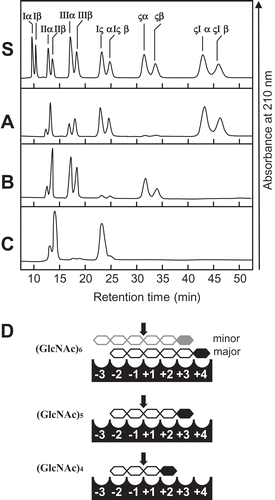
Figure 4. Time-dependent HPLC analysis of the hydrolysis products of (GlcNAc)4 by SdChiA.
The enzyme reaction was conducted for 0 min (a), 1 min (b), 5 min (c), 10 min (d) and 30 min (e), respectively.
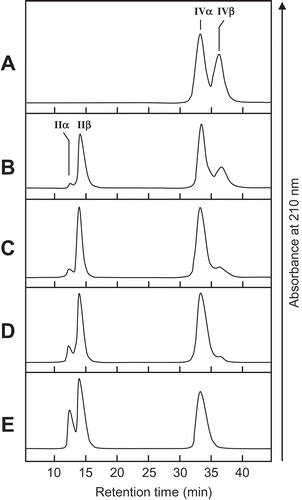
Figure 5. Cloning strategy and primary structure of SdChiA.
(a) Cloning strategy of SdChiA cDNA. Three fragments overlapped to obtain the complete nucleotide sequence of SdChiA cDNA. The open box represents the coding region and solid lines represent the 5′- and 3′-noncoding regions. The relative position and direction of each primer are indicated by arrows. The solid lines indicate cDNA fragments amplified by each PCR. SP, signal peptide. (b) Nucleotide sequence of SdChiA cDNA with its deduced amino acid sequence. The putative signal peptide at the N-terminal is indicated by a dotted underline. Internal peptide obtained by V8 protease digestion are underlined. (c) Schematic representation of SdChiA. CBD, chitin-binding domain; GH, glycoside hydrolase.
Figure 6. Alignment of amino acid sequences from domains of SdChiA with those from corresponding regions of several proteins.
(a) Alignment of sequences from chitin-binding domains of SdChiA with those of chitin-binding domain containing proteins. (b) Structure-based sequence alignments of catalytic domain among SdChiA, plant class V chitinases, human chitotriosidase, plant class III chitinases and plant class IIIb chitinase. Identical residues are shown in white and black background; dashes indicate gaps. Zigzag bold lines and arrows indicate α- and β- strands, respectively. The strands shown on the top of sequences are the secondary structures of SdChiA predicted by a computer program (PSIPRED, http://bioinf.cs.ucl.ac.uk/psipred/). The strands at the bottom of the sequences are secondary structures of hevamine. The eight β-strands and α-helixes in the TIM barrel are indicated and labeled. A box indicates a region of the putative extra α/β domain. The catalytic glutamic acid in the DXDXE motif is indicated by a triangle. SdChiA, S. doederleinii chitinase-A; CrChi-A, plant class V chitinase from Cycas revolta (accession no. BAD98525.); NtChiV, plant class V chitinase from Nicotiana tabacum (CAA54374); AtChiC, plant class V chitinase from Arabidopsis thaliana (NP_193716); Chitotriosidase, chitinase from Homo sapiens (AAG10644); Hevamine, plant class III chitinase from Hevea brasiliensis (P23472); PLChi-A, plant class III chitinase from Ananas comosus (BAG38685); PrChi-A, plant class IIIb chitinase from Pteris ryukyuensis (BAE98134).

Figure 7. Chitin-binding activity of SdChiA, GlxChi-B and CrChi-A.
(a) SDS-PAGE of three proteins (each 2 µg) applied to the chitin-binding assay. The gel was stained with CBB. Lane M, marker proteins; lane 1, SdChiA (CBM18 × 2+ GH18); lane 2, GlxChi-B (CBM18+ GH19); lane 3, CrChi-A (GH18). (b) The proteins were applied to chitin powders equilibrated with 0.15 M NaCl in 10 mM Tris-HCl. After incubation and centrifugation, the adsorbed and unadsorbed fractions were subjected to SDS-PAGE, and then stained with CBB. Lane M, maker proteins; lane N, the unadsorbed fraction; lane B, the adsorbed fraction.
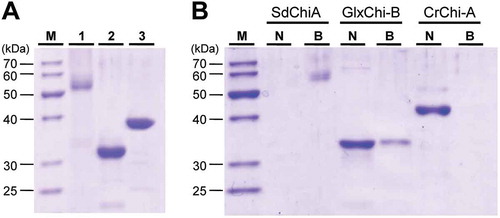
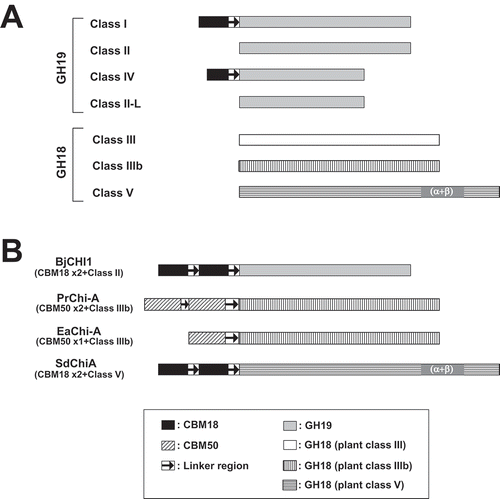
Figure 8. Classification of plant chitinases.
(a) Schematic representation of five classes (I-V) and two subclasses (II-L and IIIb) of chitinases. (b) Schematic representation of unclassified plant chitinases. BjCHI1, Brassica juncea chitinase-1 (AAF02299); PrChi-A, P. ryukyuensis chitinase-A (BAE98134); EaChi-A, E. arvense chitinase-A (BAI22848); SdChiA, S. doederleinii chitinases-A. GH and CBM indicate glycoside hydrolase and carbohydrate binding module, respectively.