Figures & data
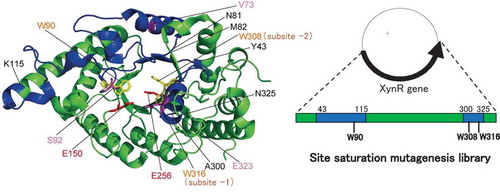
Figure 1. Amino acid sequences of xylanases.
Notes: The ClustalW program was used for multiple sequence alignment of the amino acid sequences of xylanases from Bacillus halodurans S7 (BhS7Xyn, Gene bank accession no. AY687345.1), Bacillus sp. strain TAR-1 (XynR) (JF912896.1), and Geobacillus stearothermophilus (GsXyn) (AAC98140). The asterisk indicates the amino acid residues conserved in BhS7Xyn, XynR, and XynGR40. Two catalytic Glu residues and three Trp residues conserved are boxed with a broken line. The target amino acid sequences of the site saturation mutagenesis library of XynR, Tyr43–Lys115 and Ala300–Asn325, are underlined. Val73, Ser92, and Glu323 of XynR are boxed.

Figure 2. Structures of xylanases.
Notes: The PyMOL program was used to visualize the whole structure of BhS7Xyn (PDB accession no. 2UWF) (A) and the active-site structure of GsXyn complexed with xylopentose (1R87) (B). Two catalytic Glu residues (Glu150 and Glu256) and three Trp residues (Trp90, Trp308, and Trp316) conserved are colored in red and yellow, respectively. Val73, Ser92, and Glu323 are colored in pink. The target regions to be mutated, Tyr43–Lys115 and Ala300–Asn325, are colored in blue, while other regions in green. The amino acid number indicates that corresponding to those of XynR.
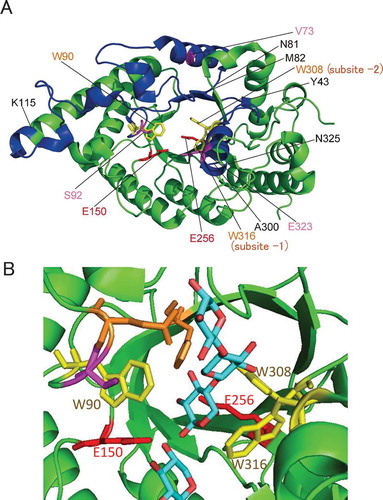
Figure 3. Sequence analysis of the site saturation mutagenesis library of XynR.
Notes: Substitution of amino acid residue is indicated by characters; deletion of one nucleotide is indicated by dot, and deletion of 11‒155 nucleotides is indicated by arrow. a‒m indicates independent clones. 93Y and 114T, to which asterisk is attached, are in the same clone.
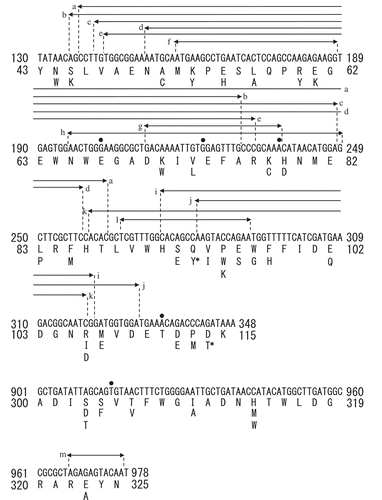
Table 1. Distribution of clones with the same activity or stability.
Figure 4. Thermostability of XynR variants.
Notes: Residual activities of WT (A), V73L, S92E, and E323A (B), and S92X (C) are shown. Hydrolysis reaction of RBB-xylan was carried out at 37ºC for 15 min with E. coli expression product of WT which received heat treatment at 60, 70 or 80ºC for 15 min (A) or V73L, S92E, and E323A (B) or S92X (C) which received heat treatment at 80ºC for 15 min. Residual activity indicates the value compared to that before heat treatment.Error bars indicate SD values for three-times measurements.
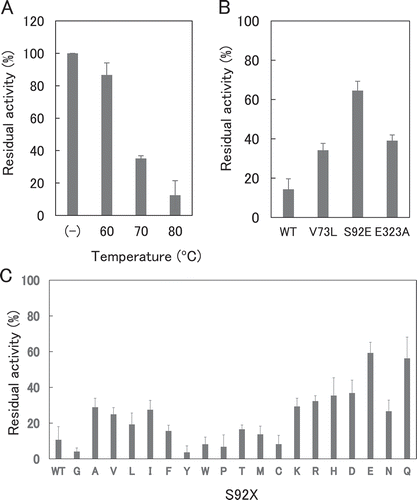
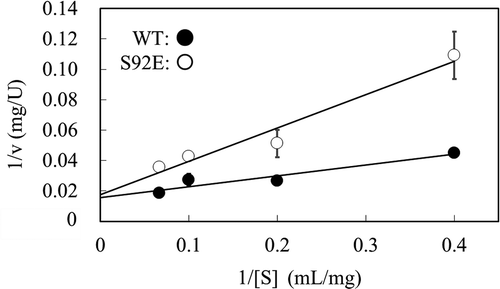
Figure 5. Steady-state kinetic analysis of thermostable XynR variant.
Notes: Hydrolysis reaction of beechwood xylan was carried out at 37ºC with the purified enzyme preparation of WT or S92E (0.1 µM). Lineweaver-Burk plot is shown. One unit is defined as the amount of enzyme that produces 1 µmol of reducing sugar per min. Error bars indicate SD values for three-times measurements.