Figures & data
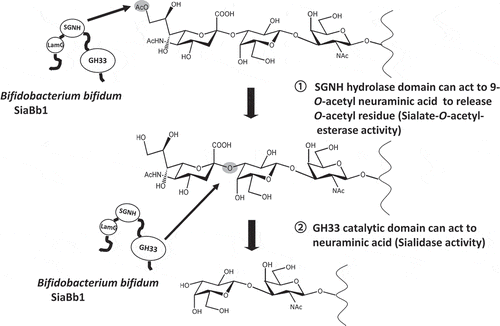
Figure 1. Schematic representation of the domain architecture of SiaBb1.
The amino acid numbering starts at the probable initiation codon, and the domain responsible for exo-α-sialidase activity (amino acids 329 to 655) is depicted as a black box. The black bars at the N- and C-termini indicate a signal peptide and a transmembrane anchor region, respectively. The schematic representation of the domain architecture of SiaBb2 is also shown (see ref.6).
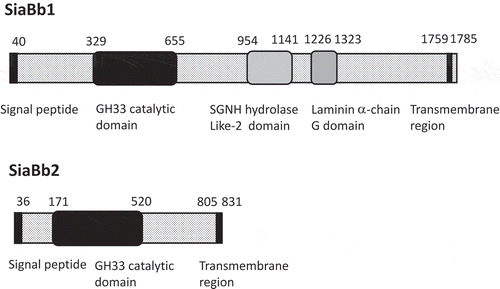
Figure 2. Comparison of the amino acid sequence of the catalytic domain of SiaBb1 with those of other structurally solved exo-α-sialidases. A multiple alignment of the catalytic domain of exo-α-sialidases was created by the ClustalW program. Similar residues, grey; identical residues, black; conserved nucleophilic tyrosine, closed arrowhead; acid/base aspartic acid, open arrowhead; BNR/Asp-box and YRIP motifs, boxed; Bb1, SiaBb1 from B.bifidum JCM1254 (AB278566); Bb2, SiaBb2 from B.bifidum JCM1254 (AB278567); Aur, exo-α-sialidase from Arthrobacter ureafaciens IFO12140 (AB193297); Jsp, exo-α-sialidase from Janibacter sp. HTCC2649 (AAMN01000001).
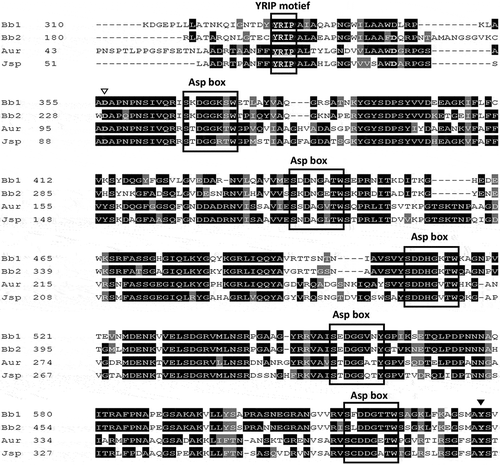
Figure 3. Analysis of the substrate specificities of SiaBb1. (A) Enzymatic reaction of SiaBb1 toward PA-3ʹ- and 6ʹ-sialyllactoses. Closed square, PA-3ʹ-sialyllactose; closed circle, PA-6ʹ-sialyllactose. (B) TLC of released sialic acids from 3ʹ- and 6ʹ-sialyllactoses by the enzymatic action of SiaBb1. TLC was developed with a solvent of 1-butanol/acetic acid/water (2/1/1) followed by spraying with the diphenylamine-aniline-phosphoric acid reagent.
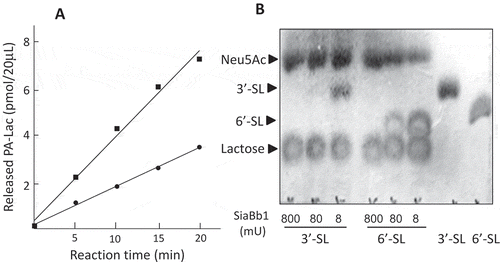
Figure 4. Multiple alignment of the amino acid sequences of the conserved region in the SGNH hydrolase-like 2 domain of SiaBb1.
A multiple alignment of the SGNH hydrolase domain from enzymes was created by the ClustalW program. Similar residues, grey; identical residues, black; conserved SGNH residues, arrowhead; acid/base aspartic acid, open arrowhead; Bb, SiaBb1 from B.bifidum JCM1254 (AB278566); Cp, lipase/acyl hydrolase from Clostridium perfringens ATCC13124 (NC008261); Pa, lypolytic enzyme from Pseudoalteromonas atlantica T6c (CP000388); Bt, lypolytic enzyme from Bacteroides thetaiotaomicron VPI-5482 (NC004663).
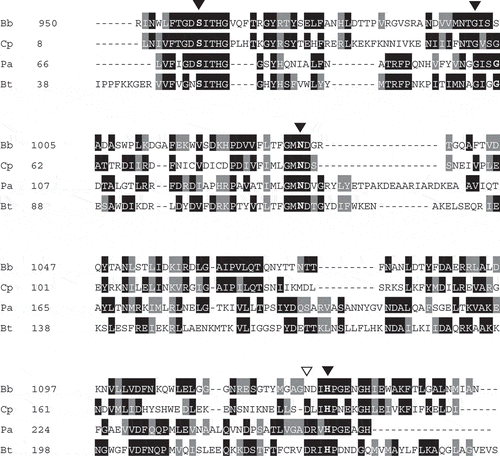
Table 1. Action of SiaBb1 to various pNP-compounds.
Figure 5. O-Acetylesterase activity of SiaBb1.
(A) Activity of SiaBb1, truncated SiaBb1 that does not contain the SGNH and Lam G domains, and SiaBb2 toward the p-NP-acetate substrate. SiaBb1, closed diamond; truncated, open square; SiaBb2, cross. (B) Actions of SiaBb1 and SiaBb2 toward Neu5,9Ac2. TLC was developed with a solvent of 1-butanol/acetic acid/water (2/1/1) followed by spraying with the diphenylamine-aniline-phosphoric acid reagent.
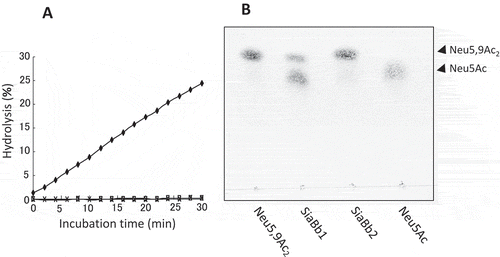
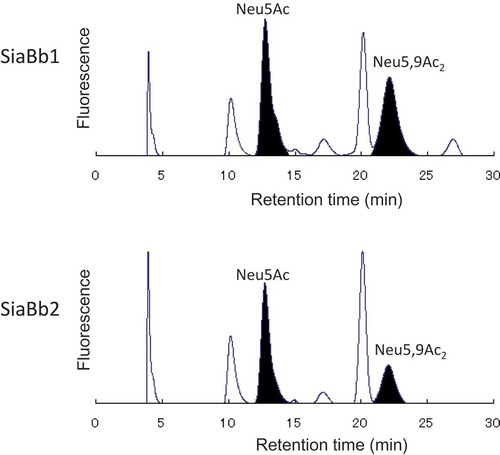
Figure 6. HPLC analysis of the released sialic acid species by the actions of SiaBb1 and SiaBb2 toward bovine submaxillary mucin.
Released sialic acid species were labeled with DMB, and analyzed by HPLC using a reversed-phase column and a fluorescence spectrophotometer. Samples were detected by excitation at 373 nm and emission at 448 nm.