Figures & data
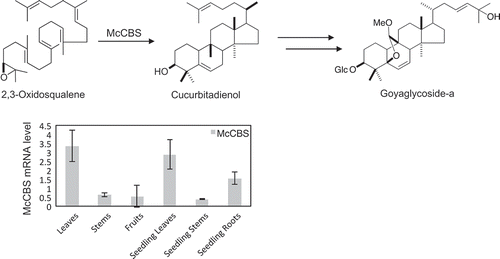
Figure 1. Biosynthetic pathway of the four types of triterpenes in M. charantia. Cucurbitadienol, isomultiflorenol, β-amyrin and cycloartenol are biosynthesized from a common precursor 2,3-oxidosqualene by four different OSCs. The subsequent modification steps involve oxidation and glycosylation.
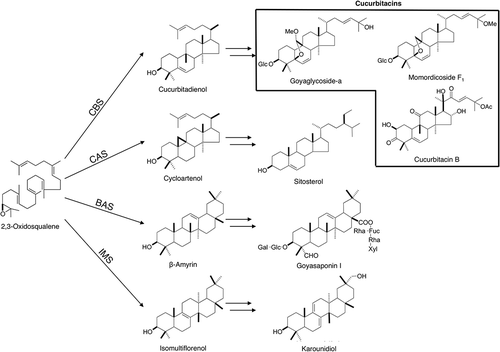
Figure 2. Phylogenetic analysis of four OSC genes from M. charantia with various plant OSC genes. The boxed genes represent McCBS, McIMS, McBAS and McCAS. Underline indicates BASs; β-amyrin synthases, and shaded areas are CASs; cycloartenol synthases, CBSs; cucurbitadienol synthases and IMSs; isomultiflorenol synthases. The detail of known OSCs is listed in Table S3.
Figure 3. Amino acid sequence alignments of McCBS, CpCPQ, McIMS and LcIMS1. Both DCTAE and MWCXC motifs are highlighted. The black dot indicates the residue two residues upstream of the DCTAE motif.

Figure 4. GC-MS chromatogram of hexane extracts from yeast transformants harboring each OSC gene. These GC-MS chromatograms are in an order from the top; hexane extracts from yeast expressing McCBS, CPQ, PSX, McCAS, McIMS, McBAS, authentic β-amyrin and empty vector. Dotted line shows OS (2,3-oxidosqualene), DOS (dioxidosqualene), Erg (ergosterol), cucurbitadienol, cycloartenol and β-amyrin.
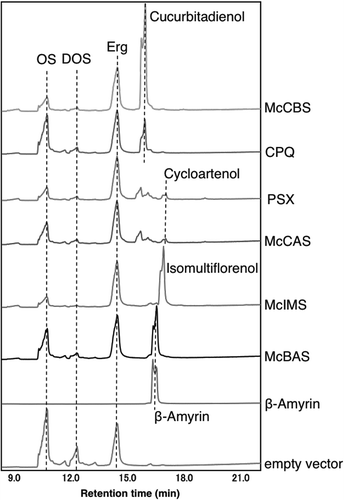
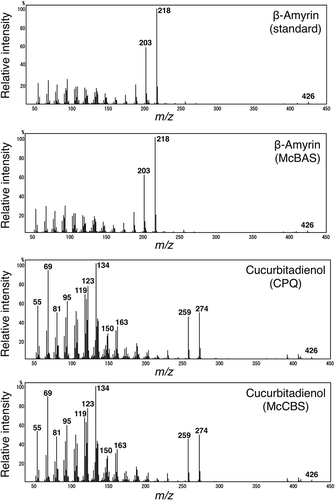
Figure 5. Mass spectra of β-amyrin or cucurbitadienol produced by McBAS or McCBS. These mass spectra are in an order from the top; authentic β-amyrin (retention time: 16.5 min, ), β-amyrin from McBAS-expressing yeast (retention time: 16.5 min, ), cucurbitadienol from CPQ-expressing yeast (retention time: 16.0 min, ) and cucurbitadienol from McCBS-expressing yeast (retention time: 16.0 min, ).