Figures & data
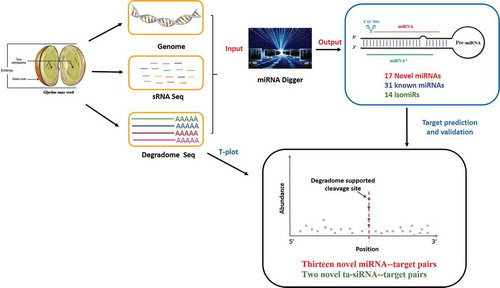
Figure 1. Workflow of miRNA Digger. miRNA Digger is free for download from http://www.bioinfolab.cn/miRNA_Digger/index.html [Citation29].
![Figure 1. Workflow of miRNA Digger. miRNA Digger is free for download from http://www.bioinfolab.cn/miRNA_Digger/index.html [Citation29].](/cms/asset/dd6c3ed7-6267-4486-951c-fd86b8ff5e28/tbbb_a_1536513_f0001_oc.jpg)
Figure 2. Novel miRNA-miRNA* duplexes discovered in soybean. A-P: The newly identified miRNAs and miRNA*s (marked with lines) were mapped to the pre-miRNAs. The degradome-supported DCL1-mediated cleavage signals were marked with arrows.
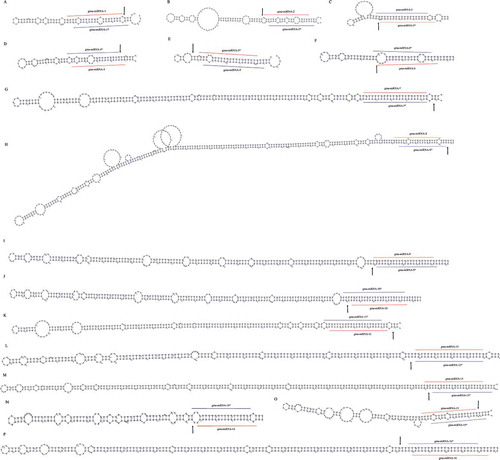
Figure 3. Novel miRNA, gam-miR482-5p.2, uncovered from known pre-miRNA. A: The newly identified gam-miR482-5p.2/gam-miR482-5p.2* (marked with lines) and miRBase registered gam-miR482-5p/gam-miR482-3p (marked with dotted lines) were mapped to the pre-miRNA. The degradome-supported DCL1-mediated cleavage signal was marked with arrow. B: Expression of gam-miR482-5p.2, gam-miR482-5p.2*, gam-miR482-5p and gam-miR482-3p in six tissues.
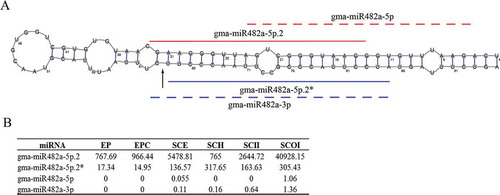
Figure 4. isomiRs uncovered in known precursors. The miRNA Digger discovered isomiRs/isomiRs* (marked with lines) and miRBase registered miRNAs counterparts (marked with dotted lines)were mapped to the pre-miRNAs. The degradome-supported DCL1-processing sites were marked with arrow.
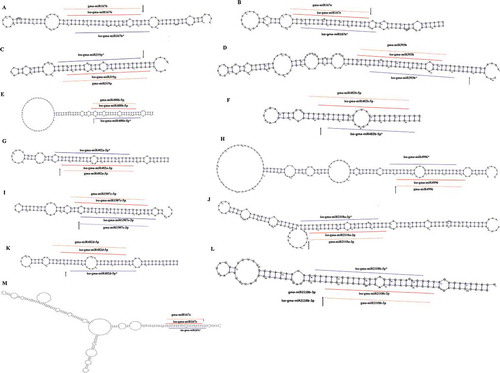
Figure 5. Expression profiling of novel miRNAs. The expression pattern of novel miRNAs in cotyledon stage across six different tissues, including: Embryo Proper (EP), Embryo Proper Cotyledon (EPC), Seed Coat Epidermis (SCE), Seed Coat Hilum (SCH), Seed Coat Inner Integument (SCII) and Seed Coat Outer Integument (SCOI) were analyzed and the log2 transformed values of abundance are displayed as a heat map.
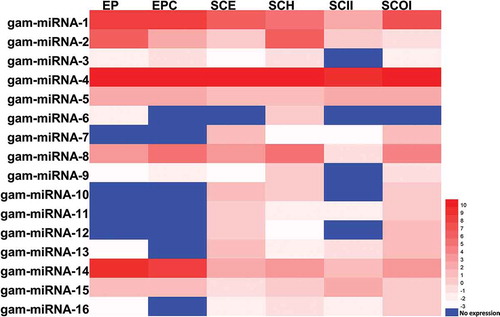
Figure 6. Expression profiling of isomiRs and their miRNA counterparts. The expression pattern of newly discovered isomiRs and their miRBase-registered mature miRNA counterparts in cotyledon stage across six different tissues, including: Embryo Proper (EP), Embryo Proper Cotyledon (EPC), Seed Coat Epidermis (SCE), Seed Coat Hilum (SCH), Seed Coat Inner Integument (SCII) and Seed Coat Outer Integument (SCOI) were analyzed and the log2 transformed values of abundance are displayed as a heat map.
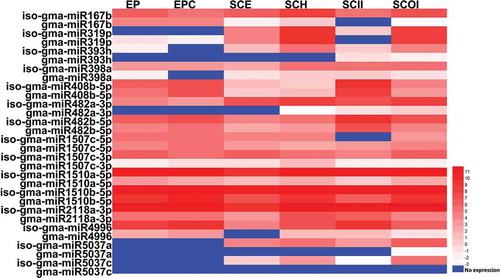
Table 1. Targets of novel miRNAs.
Figure 7. Degradome sequencing data-based validation of the miRNA-target interactions in soybean. Five libraries of degradome sequencing data libraries (GSM647200,GSM848963,GSM848964,GSM848965 and GSM848967) were recruited for T-plot profiling. The IDs of the target transcripts and the corresponding miRNAs are listed on the top. The y axes measure the normalized reads (in RMP, reads per million) of the degradome signals, and the x axes represent the position of the cleavage signals on the target transcripts. The binding sites of the phased siRNA on their target transcripts were denoted by horizontal lines, and the dominant cleavage signals were marked by arrows.
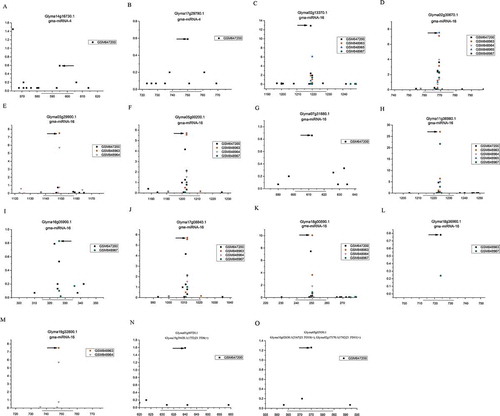
Table 2. Targets of miRNA-triggered secondary ta-siRNAs.
