Figures & data
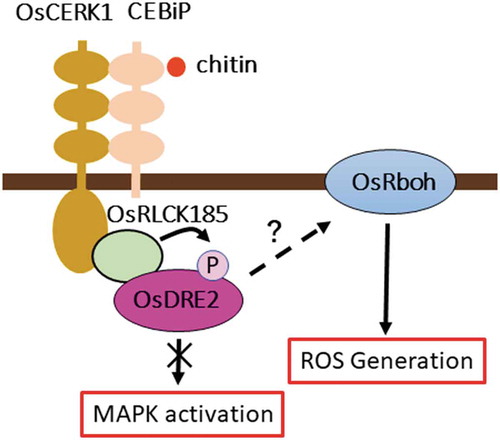
Figure 1. OsDRE2a interacts with OsRLCK185 at plasma membrane. a. Comparison of amino acid sequences between OsDRE2a and yeast Dre2.The sequence of yeast Dre2 (GeneBank accession number A7A031) was aligned with the OsDRE2a using ClustalW (http://www.ddbj.nig.ac.jp). Identical amino acid residues are indicated by black box. Shaded boxes indicate SAM methyltransferase like domain and CIAPIN1 domain, respectively. The asterisks indicate conserved cysteine residues in CIAPIN1 domain. b. OsDRE2a interacts with OsRLCK185 in yeast two-hybrid experiments. The growth of yeast colonies on plates (-ULWH) lacking uracil (U), leucine (L), tryptophan (W), and histidine (H) with 0.1 mM 3-aminotriazole (3-AT) indicates a positive interaction. c. Visualization of the interaction between OsDRE2a and OsRLCK185K108E by BiFC analysis in rice protoplasts. GUS was also used as negative control. Venus fluorescence indicates interaction between OsDRE2a and OsRLCK185K108E. Scale bar = 10µm.
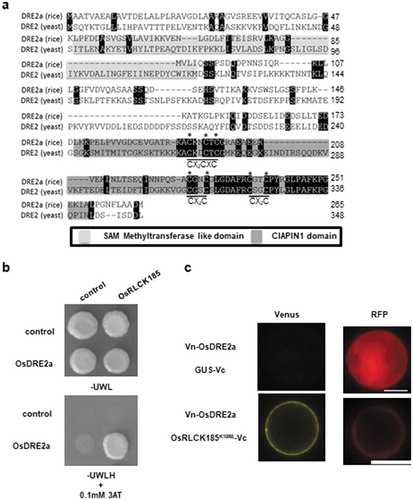
Figure 2. Conserved cysteine residues in the CIAPIN1 domain are important for the interaction between OsDRE2a and OsRLCK185. a. Schematic structures of full-length OsDRE2a (1-266aa), its N-terminus (1–132), and its C-terminus (133–266). Shaded boxes indicate SAM methyltransferase like domain and CIAPIN1 domain, respectively. The numbers indicate the positions of amino acid sequences. b. OsRLCK185 interacts with C-terminus of OsDRE2a. The full length OsDRE2a (1-266aa) was divided into N-terminus (1–132) and C-terminus (133–266). N- terminus contain the SAM methyltransferase like domain. C- terminus contains the CIAPIN1 domain. The growth of yeast colonies on plates (-ULWH) lacking uracil (U), leucine (L), tryptophan (W), and histidine (H) indicated a positive interaction. c. OsRLCK185 didn’t interact with OsDRE2aC226A/C229A in yeast two-hybrid experiment. The growth of yeast colonies on plates (-ULWH) lacking uracil (U), leucine (L), tryptophan (W), and histidine (H) indicated a positive interaction.
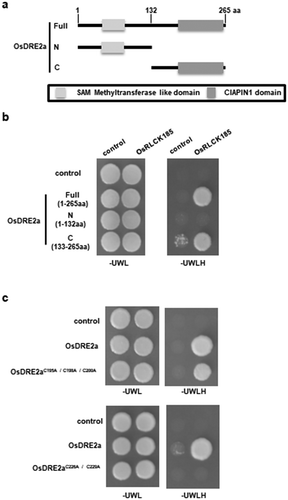
Figure 3. OsDRE2a is phosphorylated by OsRCLK185 but not OsCERK1. a. OsRLCK185 phosphorylates OsDRE2a in vitro. The in vitro phosphorylation reaction was performed using [32P]γ-ATP, and phosphorylated proteins were detected by autoradiography. CBB, Coomassie brilliant blue. b. OsCERK1 does not phosphorylate OsDRE2a in vitro. The in vitro phosphorylation reaction was performed with a recombinant of the intracellular domain of OsCERK1, kinase inactive mutant OsRLCK185K108E and OsDRE2a. The protein loading control was shown by staining with Coomassie Brilliant Blue. The in vitro phosphorylation reaction was performed using [32P]γ-ATP, and phosphorylated proteins were detected by autoradiography.
![Figure 3. OsDRE2a is phosphorylated by OsRCLK185 but not OsCERK1. a. OsRLCK185 phosphorylates OsDRE2a in vitro. The in vitro phosphorylation reaction was performed using [32P]γ-ATP, and phosphorylated proteins were detected by autoradiography. CBB, Coomassie brilliant blue. b. OsCERK1 does not phosphorylate OsDRE2a in vitro. The in vitro phosphorylation reaction was performed with a recombinant of the intracellular domain of OsCERK1, kinase inactive mutant OsRLCK185K108E and OsDRE2a. The protein loading control was shown by staining with Coomassie Brilliant Blue. The in vitro phosphorylation reaction was performed using [32P]γ-ATP, and phosphorylated proteins were detected by autoradiography.](/cms/asset/e4c14c13-97e4-4ba1-8841-323357bfbc50/tbbb_a_1543012_f0003_oc.jpg)
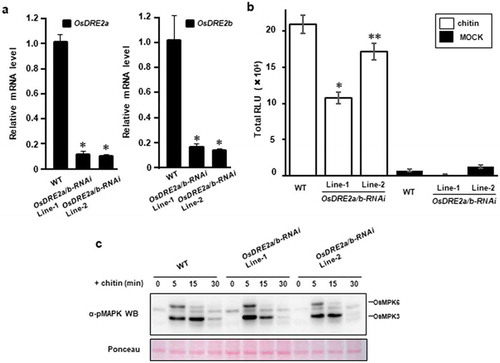
Figure 4. Two kinds of OsDRE2 proteins were conserved in rice and the expression of these genes was induced by chitin. a. Comparison of amino acid sequences between OsDRE2a and OsDRE2b.The sequence of OsDRE2b was aligned with that of OsDRE2a using ClustalW. Identical amino acid residues are indicated by black box. b. Subcellular localization of OsDRE2a and OsDRE2b. OsDRE2a-GFP and OsDRE2b-GFP were expressed under the control of 35S promoter in rice protoplasts. Scale bar = 10µm. c. Chitin-induced expression of OsDRE2a and OsDRE2b. The expression of OsDRE2a and OsDRE2b was analyzed by real time RT-PCR using specific primers. Ubiquitin (Ubq) was used as an internal control. Data are means and SDs. The asterisks indicate significant differences from the normal condition (0h) controls by Student’s t-test (*P < 0.01).
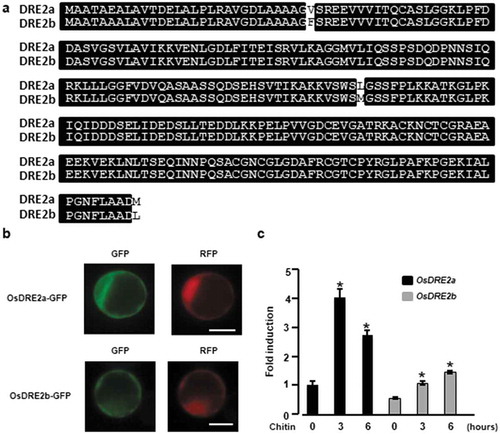
Figure 5. OsDRE2a and OsDRE2b positively regulate ROS burst. a. Transcript levels of OsDRE2a and OsDRE2b in the OsDRE2a/b RNA interference (RNAi) suspension cells under normal growth condition were determined by quantitative real-time PCR using specific primers. Ubiquitin (Ubq) was used as an internal control. Data are means and SDs. The asterisks indicate significant differences from the wild-type (WT) controls by Student’s t-test (P < 0.01). b. Production of ROS in OsDRE2a/b RNAi cells after chitin treatment. Rice suspension cells were treated with 2 μg/mL chitin in a solution containing 500 μM L-012 and 10 μg/mL horseradish peroxidase. Total relative light units (RLU) during 50 min of treatment are presented to indicate the ROS production. Data are means and SEs (n = 9; Chitin treatment, N = 3; Mock treatment). The asterisks indicate significant differences from the wild-type (WT) controls by Student’s t-test (*P < 0.01, **< 0.05). c. Chitin -induced activation of mitogen-activated protein kinase (MAPK) in OsDRE2a/b RNAi cells. Total protein was extracted from rice suspention cells treated with 2 μg/mL chitin. The activation of MAPK was analyzed by immunoblot with anti-pMAPK antibody. The protein loading control was shown by Ponceau staining. Experiments were repeated at least three times with similar results.