Figures & data
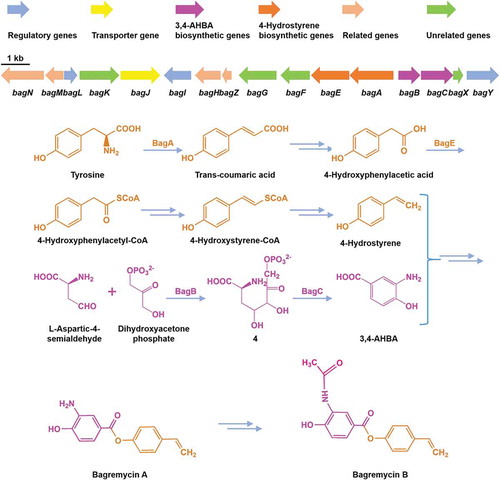
Table 1. Strains and plasmids used in this study.
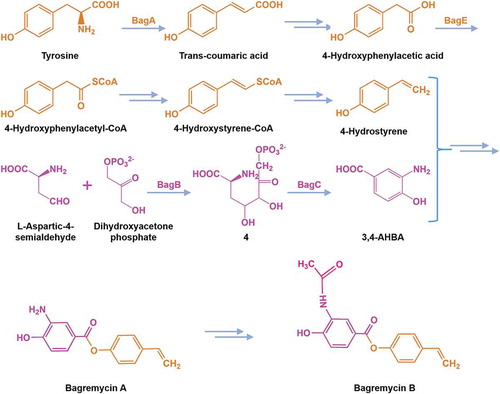
Figure 1. Genetic organization of bagremycins biosynthetic gene cluster (BGC). The proposed BGC contains 11 putative biosynthetic and regulatory genes, 1 resistance gene and 4 unrelated genes.
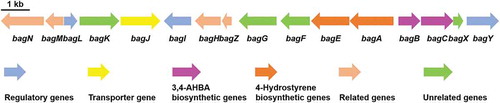
Table 2. Proposed functions of ORFs within the bagremycins biosynthetic gene cluster.
Figure 2. Construction of bagE disruption mutant TDRE. (a) Schematic representation of bagE disruption. The internal region of bagE was replaced by neomycin resistance gene (neor). (b) Identification of bagE mutant TDRE by PCR. Line M indicated the DNA molecular weight marker. Line U and D indicated PCR products of the fragments containing the bagE-upstream and bagE-downstream regions, respectively. 1, 2 and 3 represent three conjugants selected by resistant screening and are used for PCR identification. c Transcriptional analysis of bagE and its flanking genes in the wild-type strain S. sp. Tü 4128 (+) and the bagE mutant TDRE. 16S rRNA was used for internal control. – indicated the negative control.
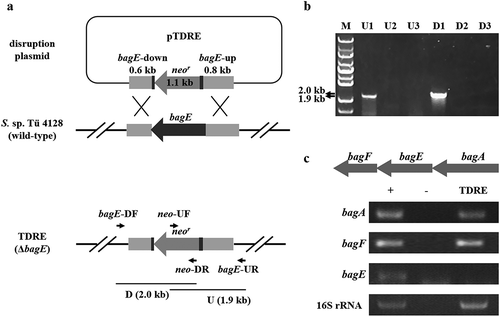
Table 3. Analysis for bagremycins productions of relative genes disruption mutants.
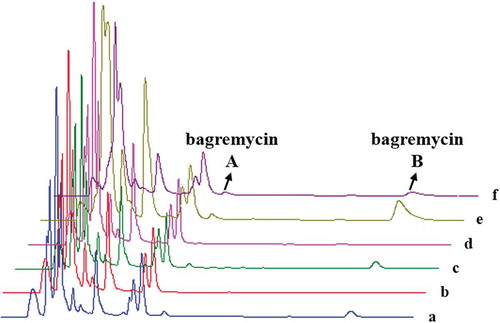
Figure 3. HPLC chromatograms of organic extracts of wild-type strain S. sp. Tü 4128 (a), bagE mutant TDRE (b), bagE complementation strain TCPE (c), bagE complementation control (d), bagE overexpression strain TOEE (e), bagE overexpression control (f).