Figures & data
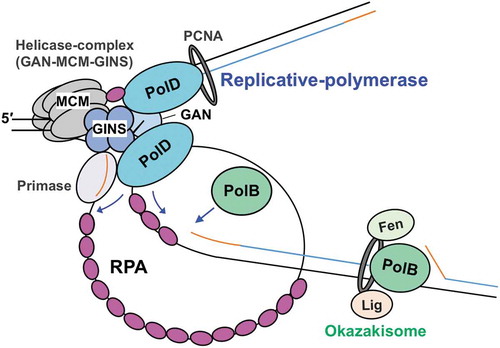
Figure 1. Preparation of RPA complex from Thermococcus kodakarensis (TkoRPA). (a) Schematic map of the gene organization at the operon of rpa41-rpa14-rpa32 in the T. kodakarensis genome. (b) Purified recombinant TkoRPA14 (0.5 μg, lane 1), TkoRPA41 (0.5 µg, lane 2), and TkoRPA complex (2 μg, lane 4) proteins were subjected to SDS −17.5% PAGE followed by CBB staining. Protein size markers were run in lane 3, and their sizes are indicated on the left of the gels. (c) Recombinant RPA complex (20 fmol, lane 1, indicating “r” at the top of the panel) and the cell extracts (1.6 × 105, 8 × 105, 4 × 106, and 2 × 107 cells in lanes 2–5) were loaded to SDS-17.5% PAGE, followed by western blot analysis. The anti-PfuRPA32 antiserum was used to detect TkoRPA32 in the TkoRPA complex or in the cells. Asterisk (*) may indicate the degradation product of TkoRPA32 in the cell. (d) The properties of the purified TkoRPA41 (lane 1), TkoRPA14 (lane 2), and TkoRPA complex (lane 3) in 8% native PAGE run in TBE followed by CBB staining (left of the panel) and western blot analysis (right of the panel), using anti-PfuRPA32 antiserum.
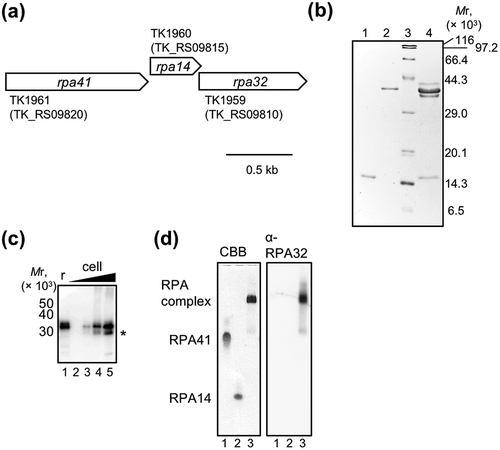
Figure 2. DNA-binding properties of TkoRPA.
Electrophoretic mobility shift assay (EMSA) of TkoRPA complex, TkoRPA41, and TkoRPA14 were performed with 5 nM Cy5-labeled ssDNA (a) and dsDNA (b). DNA and various concentrations of TkoRPAs were incubated at 50°C for 15 min. The band assignments are indicated on the side of the panels.
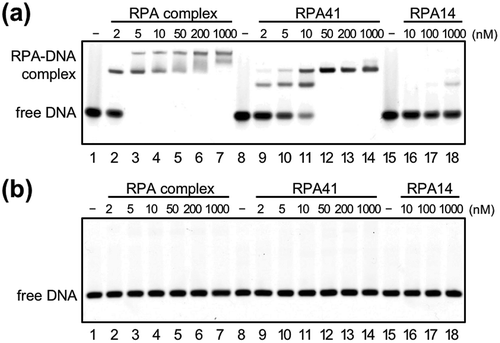
Figure 3. Effects of TkoRPA complex on DNA synthesis reactions by TkoPolB and TkoPolD.
(a) The primer extension reactions were performed with M13mp18 ssDNA as the template, in the presence or absence of TkoRPA complex. The reaction products from a 5′-Cy5-labeled primer were fractionated by 1.5% alkaline agarose gel electrophoresis and visualized with the Typhoon Trio+ imager. Lane 13 shows the size marker DNAs and the sizes indicated on the right of the panel. The DNA synthesis arrested point was indicated by asterisk(*) on the left. (b) The same gel electrophoresis with different contrast of the panel (a) is shown.
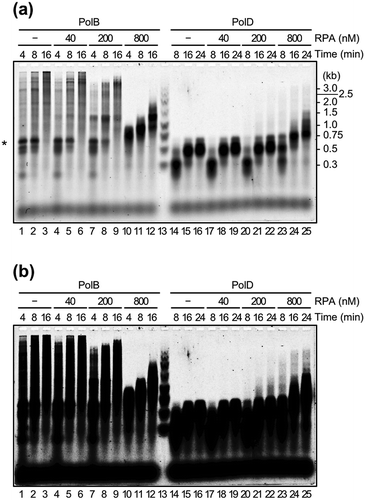
Figure 4. TkoRPA complex interacts directly with TkoPolD, TkoPolB, and various proteins.
(a) EMSA of TkoRPA complex and various proteins were performed in 5% native PAGE followed by CBB staining. Purified TkoPolD, TkoPrimase, TkoPolB, TkoRadA, TkoMCM, TkoGINS, and TkoGAN were incubated with TkoRPA complex at 50°C for 5 min. (b) EMSA of TkoRPA complex (10 pmol), TkoPolB (2, 20 pmol), and TkoPolD (2, 20 pmol) to 10 nM Cy5 labeled ssDNA were performed in 1% AGE in 0.5 × TBE. −, +, and ++ stand for 0, 2, and 20 pmol of polymerases, respectively.
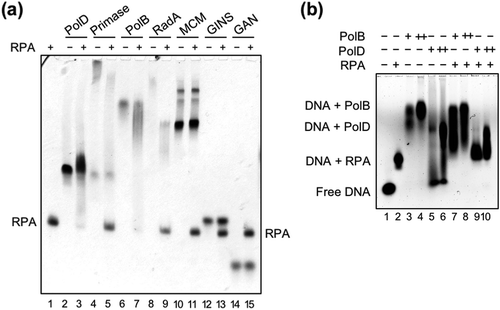
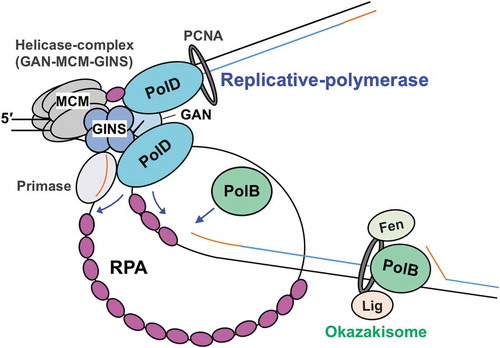
Figure 5. A model for the function of RPA in the progression of T. kodakarensis DNA replication.
The replicative helicase complex (consisting of GAN, MCM, and GINS) unwinds template DNA, generating ssDNA that is coated by RPA. RPA also contributes to the progression of the replication fork by unfolding the ssDNA secondary structure. Both PolB and PolD can progress DNA synthesis while removing RPA. Template DNA, nascent DNA, and primer are shown as black, blue, and orange lines, respectively.