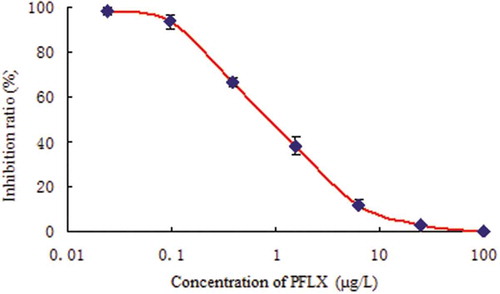
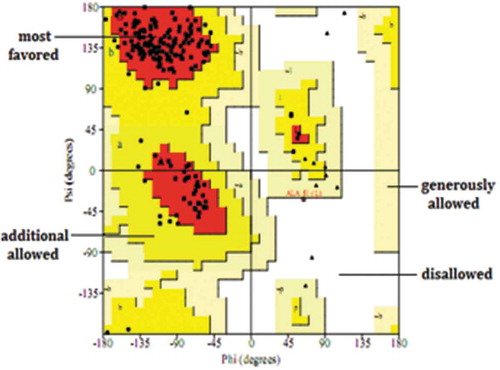
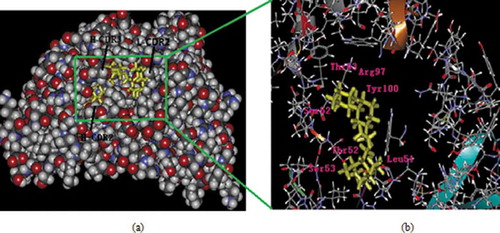
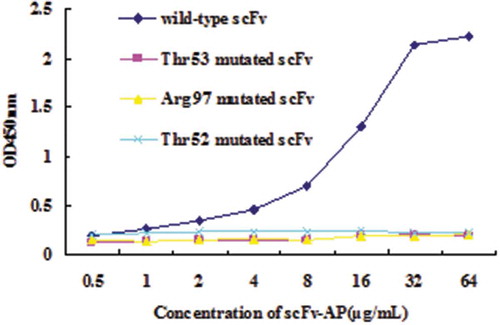
Figures & data
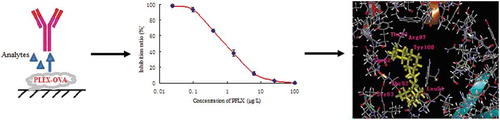
Table 1. Cross reactions (CRs) of mAb with PFLX and other bactericide analogs.
Figure 2. Cloning of the VH and VL chain genes and the scFv gene assembly. (a) Agarose gel electrophoresis of VH and VL genes of PFLX. Lane M, DL 2000 DNA marker; Lane 1, VL gene. Lane 2, VH gene. (b) Agarose gel electrophoresis of scFv of PFLX. Lane M, DL 2000 DNA marker; Lane 1, scFv gene.
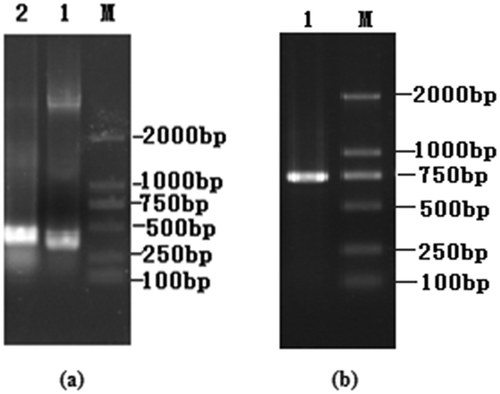
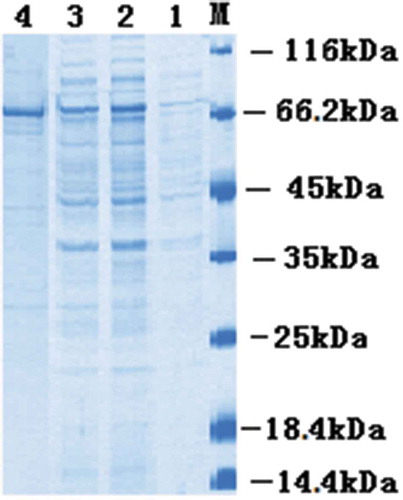
Figure 3. SDS–PAGE analysis of the anti–PFLX scFv–AP fusion protein.
Lane M, marker 116 kDa; Lane 1, before induction; Lane 2, after induction; Lane 3, the supernatant; Lane 4, purified scFv-AP.