Figures & data
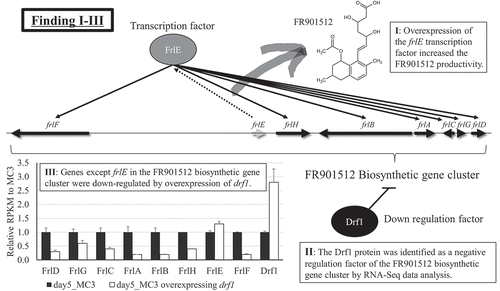
Figure 1. Productivity of FR901512 with overexpressing genes of transcription factor, FrlE, and global regulators, LaeA and VeA.
Productivity is shown as the amount of FR901512 produced per dry cell weight of each overexpression mutant relative to the MC3 strain. Three replicate experiments were performed, and the values of the means and standard deviations are presented (**, p < 0.01 [Student’s t test]).
![Figure 1. Productivity of FR901512 with overexpressing genes of transcription factor, FrlE, and global regulators, LaeA and VeA.Productivity is shown as the amount of FR901512 produced per dry cell weight of each overexpression mutant relative to the MC3 strain. Three replicate experiments were performed, and the values of the means and standard deviations are presented (**, p < 0.01 [Student’s t test]).](/cms/asset/2ae24c7e-6772-49f6-8c58-e1618d3855bb/tbbb_a_1584519_f0001_b.gif)
Figure 2. Results of qRT-PCR analysis. Relative expression levels of genes of (a) frlE, (b) laeA, (c) veA, and (d) polyketide synthase gene frlB on day 5 are shown.
Three strains (MC3, Nos. 1019 and 1407) described as “Orig.” were used as hosts to overexpress the genes encoding FrlE, LaeA, and VeA. In respective strains, expression level of each gene was compensated by that of actin gene, and then relative expression level was calculated. Three replicate experiments were performed, and the values of the means and standard deviations are presented. As another version of this in different description, the relative expression levels of respective genes in each mutant to “Orig.” of the MC3 strain are shown in Figure S4.
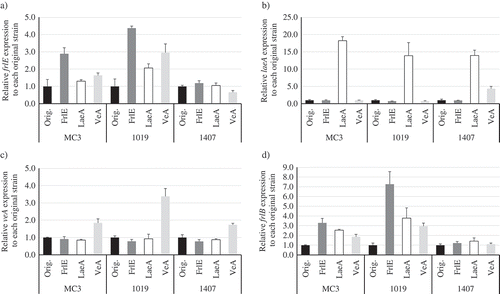
Table 1. Transcription factors correlated with the expression level of frlE (top five genes).
Figure 3. Results of RNA-Seq data analysis. RPKMs of a) frlE and b) frlB in the wild-type MC3 and the derivative four UV mutant (Nos. 425, 1019, 1225 and 1407) strains are shown.
Three replicate experiments were performed, and the values of the means and standard deviations are presented.
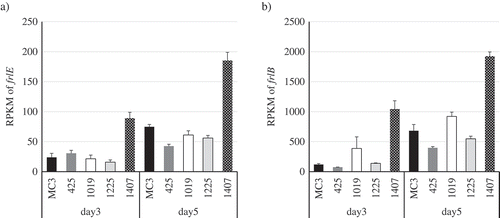
Figure 4. Productivity of FR901512 with overexpressing drf1 gene encoding Drf1.
The wild-type strain MC3 described as “Orig.” was used as a host for overexpression. Three replicate experiments were performed, and the values of the means and standard deviations are presented (**, p < 0.01 [Student’s t test]).
![Figure 4. Productivity of FR901512 with overexpressing drf1 gene encoding Drf1.The wild-type strain MC3 described as “Orig.” was used as a host for overexpression. Three replicate experiments were performed, and the values of the means and standard deviations are presented (**, p < 0.01 [Student’s t test]).](/cms/asset/291f4707-1fcb-4459-8ab3-5fcbe61579d3/tbbb_a_1584519_f0004_b.gif)
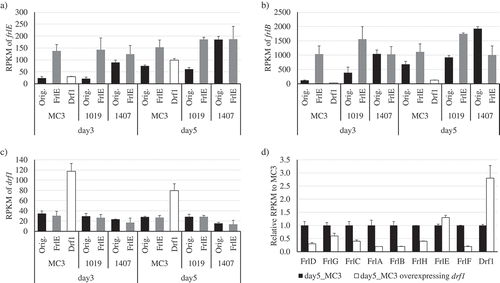
Figure 5. Results of RNA-Seq data analysis. RPKMs of (a) frlE, (b) frlB, and (c) drf1 are shown. Three strains (MC3, Nos. 1019 and 1407) described as “Orig.” were used as hosts to overexpress the genes encoding FrlE, LaeA, and VeA. (d) Effect of drf1 overexpression on the expression of genes in FR901512 biosynthetic gene cluster are shown. The genes shown encode FrlD, Transesterase; FrlG, Thioesterase; FrlC, Enoyl reductase; FrlA, P450 monooxygenase; FrlB, Polyketide synthase; FrlH, HMG-CoA reductase; FrlE, Transcription factor; and FrlF, Polyketide synthase. Three replicate experiments were performed, and the values of the means and standard deviations are presented.