Figures & data
Figure 1. Schematic diagram illustrating the procedure for isolation of temperature-sensitive mutants.
Randomly mutagenized PCR fragments of GOI containing a drug resistance marker gene are transformed into a wild type fission yeast strain (a-b). Cells are allowed to grow on non-selective medium overnight for endogenous gene replacement to occur. Transformants are then selected on plates containing the selection drug at 27°C for 3 to 4 days (c). Replicated plates are incubated at 27°C and 36°C. Temperature sensitive mutants are those that do not grow at 36°C, but grow normally at 27°C. Phloxine B (PhB) is used to help detect dead cells, which are stained dark red with this dye. Candidate ts colonies are backcrossed with a wild type strain to examine co-segregation between the ts phenotype and drug resistance (d). Spot tests are performed at various temperatures to determine the restrictive temperature (e.g. 33°C and 36°C, (e)).
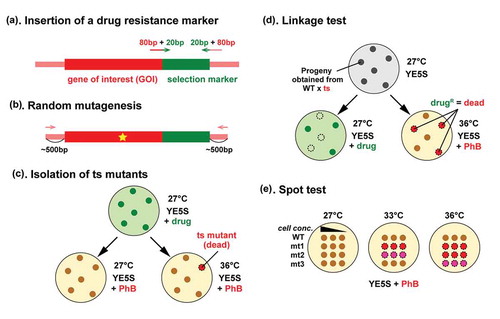
Figure 2. Spot tests and mutated amino acid residues within individual pcp1 mutants (a). Spot test. 5 × 104 cells of individual strains were spotted on the left-most line on YE5S plates and ten-fold serial dilution assays were performed. Plates were incubated at 27°C or 36°C for 3 days. (b). Summary of mutation sites in the pcp1 ts mutants. Functional domains within Pcp1 (e.g. SPM, CM1, and PACT) [Citation10,Citation15] are indicated together with mutated amino acid residues in individual ts mutants. SPM, CM1, and PACT stand for Spc110/Pcp1 Motif, Centrosomin Motif 1, and Pericentrin-AKAP450 Centrosomal Targeting, respectively.
![Figure 2. Spot tests and mutated amino acid residues within individual pcp1 mutants (a). Spot test. 5 × 104 cells of individual strains were spotted on the left-most line on YE5S plates and ten-fold serial dilution assays were performed. Plates were incubated at 27°C or 36°C for 3 days. (b). Summary of mutation sites in the pcp1 ts mutants. Functional domains within Pcp1 (e.g. SPM, CM1, and PACT) [Citation10,Citation15] are indicated together with mutated amino acid residues in individual ts mutants. SPM, CM1, and PACT stand for Spc110/Pcp1 Motif, Centrosomin Motif 1, and Pericentrin-AKAP450 Centrosomal Targeting, respectively.](/cms/asset/2d2dc175-ef47-4a1b-8b46-f8f8d8f92280/tbbb_a_1611414_f0002_oc.jpg)
