Figures & data

Table 1. Strains and plasmids used in this study.
Figure 1. Construction of plasmids for the promoter assay.
rep: replication-related gene, mob: mobilization-related gene, oriV: replication origin, Cmr: chloramphenicol resistance gene, MluI and SpeI: restriction enzyme sites.
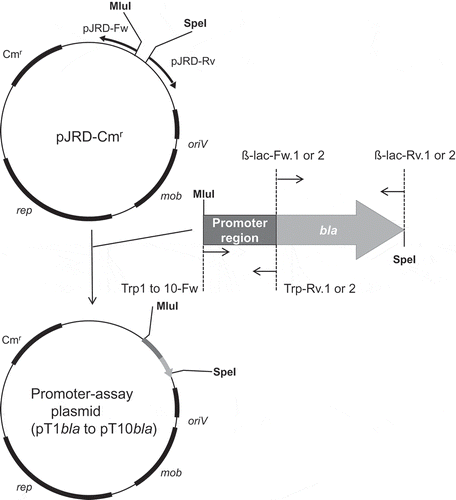
Figure 2. The trp operon of S. livingstonensis Ac10 and the 5ʹ-untranslated region (5ʹ-UTR) of trpE tested for its promoter activity.
(a) Genetic region containing the trp operon of S. livingstonensis Ac10. The scale bar represents 1 kb. (b) Schematic illustration of 5ʹ-UTR of trpE tested for its promoter activity by using the BLA gene as a reporter. DNA fragments of the 5ʹ-UTR of trpE (T1–T10) were fused with the BLA-coding gene and introduced into the promoter-assay plasmid as shown in . Putative promoter sequences were predicted by Neural Network Promoter Prediction (ver.2.2) (http://www.fruitfly.org/seq_tools/promoter.html), and the locations of the sequences with a prediction score over 0.95 are indicated with triangles.
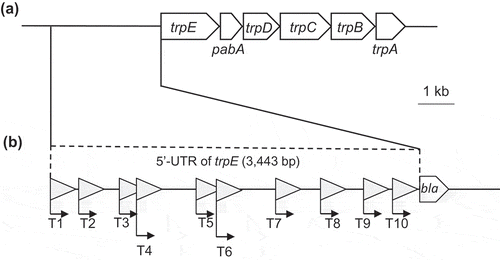
Figure 3. Effects of L-Trp supplementation on the growth of S. livingstonensis Ac10-Rifr and expression of its Trp-biosynthesis genes.
(a, b) Growth curves of S. livingstonensis Ac10-Rifr. The cells were grown at 18°C (a) and 4°C (b) in modified DSMZ medium 79 with 0.5% w/v casamino acids supplementation containing (■) and absent of L-Trp (□) at a final concentration of 0.1%. The statistical analysis was performed using a Student’s t-test from three independent experiments. Error bars indicate SD. An asterisk (*) indicates a statistically significant difference (Student’s t-test, P < 0.01). (c, d) Transcription levels of the Trp-biosynthesis genes. The ratios of the amounts of mRNA of the genes in the trp operon in the cells grown with or without 0.1% L-Trp are shown. The cells were grown at 18°C (c) or 4°C (d) and harvested in the log phase. All values were normalized to the amounts of 16S rRNA. Error bars represent SD from three independent experiments.
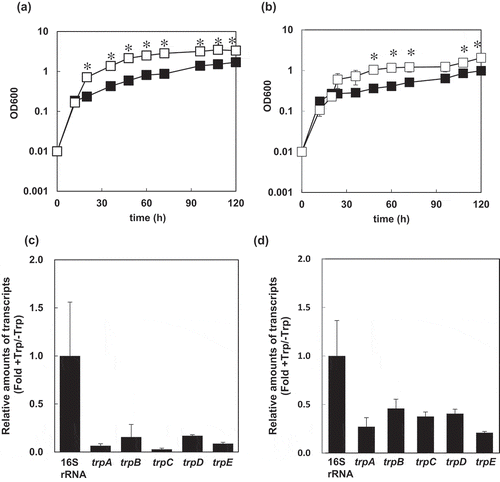
Figure 4. Production of BLA by recombinant S. livingstonensis Ac10-Rifr harboring various promoter-assay plasmids.
Specific activities of BLA produced by recombinant S. livingstonensis Ac10-Rifr cells (BLAT 1 to BLAT10) harboring the promoter-assay plasmids, pT1bla to pT10bla, respectively, are shown. BLAT0 is a negative control strain harboring pbla, which contains the BLA gene without a promoter sequence. The cells were grown at 18°C (a) and 4°C (b) in the presence (open bars) or absence (closed bars) of 0.1% L-Trp and harvested in the stationary phase (OD600 = 1.8–2.0). Error bars represent SD from three independent experiments. An asterisk (*) indicates a statistically significant difference (Student’s t-test, P < 0.01).
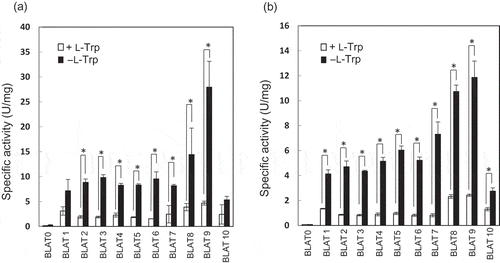
Figure 5. Expression of bla in recombinant S. livingstonensis Ac10-Rifr harboring pT9bla (BLAT9) .
(a, b) SDS-PAGE analysis of proteins of BLAT9 grown with or without L-Trp at 18°C (a) and 4°C (b). Arrowheads indicate the position of the BLA protein band. M: protein marker; Cont.: BLAT0 grown without L-Trp. (c, d) Relative amounts of bla mRNA in BLAT9 grown with 0.1% L-Trp in comparison with that in the cells grown without L-Trp. The cells were grown at 18°C (c) and 4°C (d). All values were normalized to the amounts of 16S rRNA. Error bars represent SD from three independent experiments.
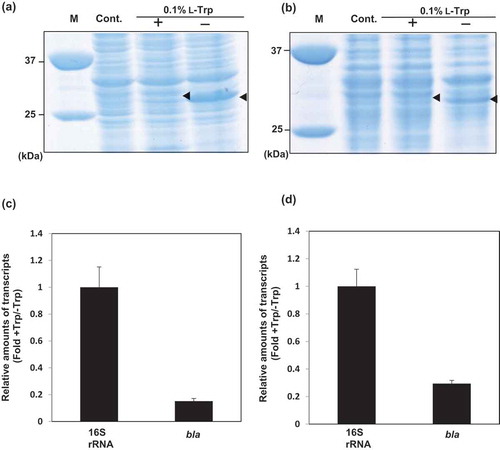
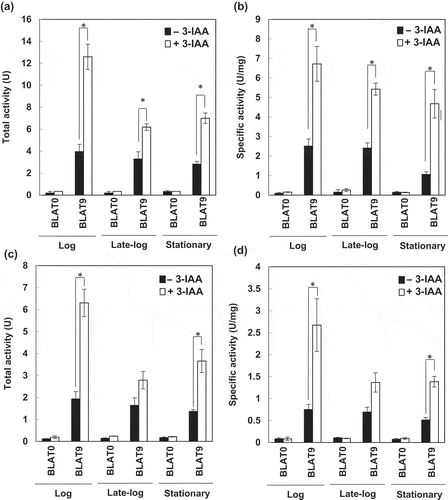
Figure 6. Effects of 3-indoleacrylic acid (3-IAA) on expression of the gene under the control of the T9 promoter.
Total activity (a and c) and specific activity (b and d) of BLA from a 5 mL culture of BLAT9. The cells grown at 18°C (a and b) and 4°C (c and d) to the log (OD600 = 0.7–0.8), late-log (OD600 = 0.8–1.0), and stationary (OD600 = 1.8–2.0) phases were transferred to LB medium with (open bars) or without (closed bars) 0.1% 3-IAA supplementation. As a control, BLAT0 harboring pbla, which contains the BLA gene without a promoter, was used. Error bars represent SD values from three independent experiments. Asterisks (*) indicate statistically significant differences (Student’s t-test, P < 0.01).