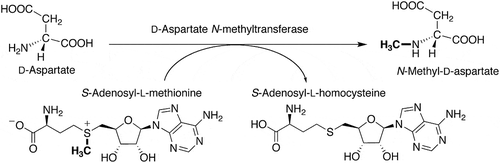
Figures & data
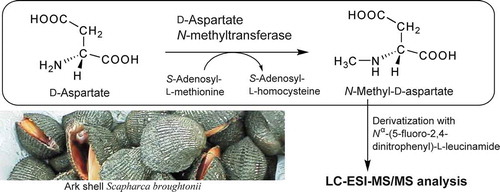
Figure 2. ESI-MS and MS/MS spectra of FDLA derivatives of authentic N-methyl amino acids.
(a) ESI-MS spectrum of FDLA derivatives of authentic NMA. (b) ESI-MS spectrum of FDLA derivatives of authentic NMG. (c) MS/MS spectrum for the precursor ion at m/z 442 shown in a. (d) MS/MS spectrum for the precursor ion at m/z 456 shown in b.
Table 1. Validation parameters for NMDA, NMLA, NMDG, and NMLG.
Figure 3. LC-ESI-MS/MS chromatograms of the FDLA derivatives of authentic N-methyl amino acids and from the assay solutions for D-aspartate N-methyltransferase in S. broughtonii.
(a) LC-ESI-MS/MS chromatograms of the FDLA derivatives of authentic NMDA and NMLA (solid line) with m/z 288 as the monitoring ion, and authentic NMDG and NMLG (dashed line) with m/z 280 as the monitoring ion. A mixture of 5 µmol/L each of NMDA, NMLA, NMDG, and NMLG was derivatized with FDLA.(b) LC-ESI-MS/MS chromatograms of FDLA derivatives in the assay solution prepared from the dialyzed tissue homogenate from the mantle of S.broughtonii, D-aspartate, and SAM without incubation at 30°C (blank). The monitoring ions used the fragment ion of m/z 288 produced from the precursor ion of m/z 442.(c) LC-ESI-MS/MS chromatograms of FDLA derivatives from the assay solution, prepared from the same dialyzed tissue homogenate used in (b), D-aspartate, and SAM with incubation for 10 min at 30°C. The monitoring ions used the fragment ion of m/z 288 produced from the precursor ion of m/z 442.
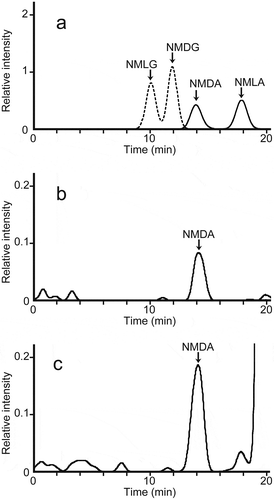
Table 2. D-aspartate N-methyltransferase activity in the tissues of S. broughtonii.