Figures & data
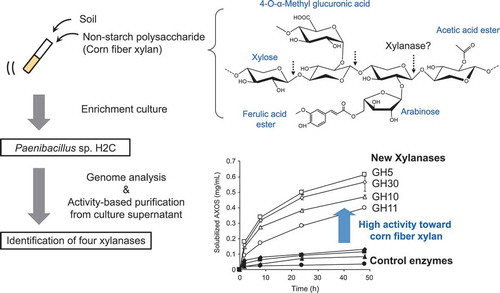
Table 1. Sugar composition (wt%) of substrate derived from corn.
Figure 1. Purification of corn fibre xylan solubilizing enzymes from culture broth of strain H2C.
(a) Relative activity of each fraction obtained by hydrophobic interaction chromatography. The activities were determined by quantification of solubilized AXOS from Cellfer after reaction. The value of the highest activity was defined as 100%. The fractions pooled and subjected to subsequent purification steps are indicated as fr.1 and fr. 2. (b) SDS-PAGE of purified fractions. Lane M: molecular mass standards; lanes 1A, 1B, and 2A: purified fractions. The SDS-PAGE gels were stained with SYPRO Ruby protein gel stain (Thermo Fisher Scientific).
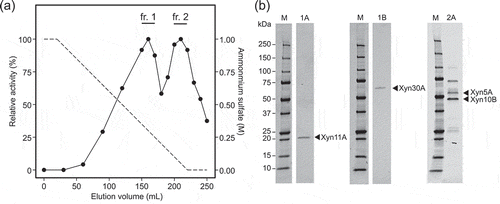
Table 2. Primers used for cloning.
Table 3. Identification of proteins in active fractions.
Figure 2. Module organizations of Paenibacillus sp. H2C xylanases Xyn5A, Xyn10B, Xyn11A, and Xyn30A.
The size of each enzyme and module is scaled according to the polypeptide chain length. GH, glycoside hydrolase family; CBM, carbohydrate binding module family. a.a., amino acids.
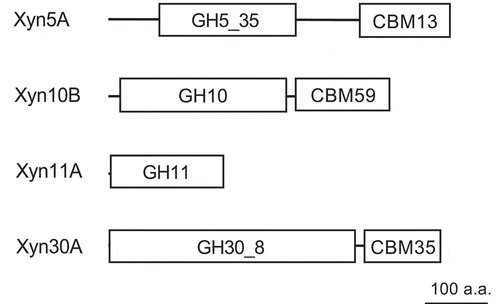
Figure 3. Phylogenetic tree of GH5_35 and GH5_21 enzymes based on maximum likelihood with 100 bootstrap replications.
Xyn5A from Paenibacillus sp. H2C is marked with a star. Bootstrap values <50 are not shown.
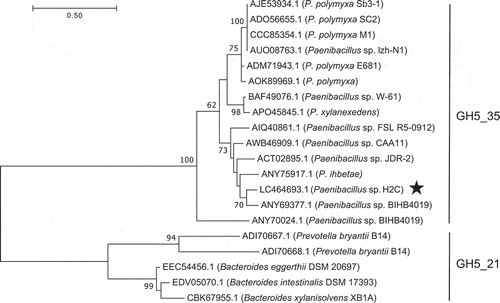
Table 4. Degradation activities toward BWX, WAX, and DDGS.
Figure 4. Time course of hydrolysis of corn dried distiller’s grains with solubles (DDGS) by 1 µg/mL of enzymes.
Purified enzymes were incubated with 2.5% DDGS in 50 mM Britton-Robinson buffer, pH 6.0. Concentrations of AXOS in soluble fraction was measured. Error bars represent SDs for duplicate reactions, each measured once.
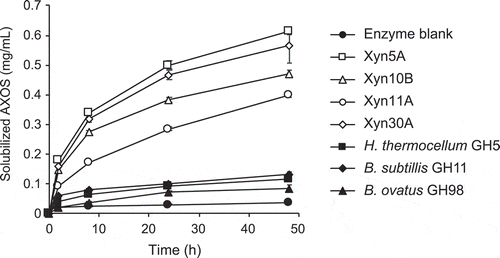
Table 5. Arabinose to xylose ratio of AXOS solubilized from DDGS.
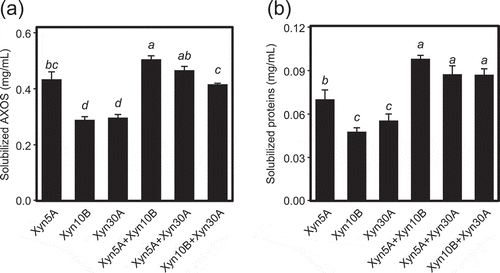
Figure 5. Combinatorial effects of xylanases on release of AXOS (a) and proteins (b) from corn dried distiller’s grains with solubles (DDGS).
Ten µg/mL of a single enzyme or a mixture of 5 µg/mL each of two xylanases, for a total of 10 µg/mL of enzyme, was incubated with 2.5% (w/v) DDGS in 50 mM Britton-Robinson buffer, pH 6.0 for 5 h. The concentration of solubilized AXOS and protein were calculated by subtracting the value of the enzyme blank. Error bars represent SDs for triplicate reactions, each measured once. Bars with identical letters indicate no significant difference from one another by the Tukey test at the 5% probability level.