Figures & data
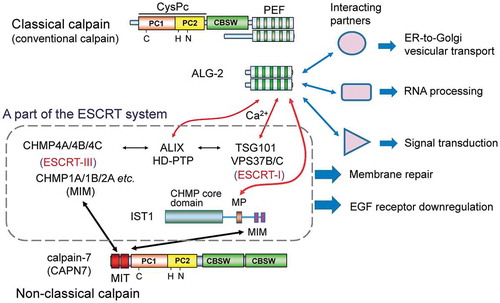
Figure 1. Relationship between the penta-EF-hand (PEF) protein family and the calpain family in mammals.
Classical (typical) calpain sequences contain the PEF domain and the calpain type β-sandwich domain (CBSW) in addition to the cysteine protease core domain (CysPc), which is further divided into two subdomains named PC1 (containing a catalytic Cys residue) and PC2 (containing catalytic His and Asn residues). Conventional calpains (μ-calpain and m-calpain) are comprised of each catalytic large subunit (designated CAPN1 for μ-calpain or CAPN2 for m-calpain) and a common regulatory small subunit (CAPNS1). Non-classical (atypical) calpain sequences lack the PEF domain but contain additional domains or motifs [calcium-binding C2 domain, microtubule-interacting and trafficking (MIT) domain, Zinc finger (ZnF), SOL-homology domain (SOH), and circularly permutated globin domain (cpGB) split by the calmodulin-binding IQ motif]. Calpain-3 (CAPN3), specifically expressed in skeletal muscles, has distinct sequences: N-terminal sequence (NS), insertion sequence 1 (IS1), and insertion sequence 2 (IS2). Calpain-7 (CAPN7) is an ortholog of fungal PalB. PEF proteins are classified into two groups based on similarity of the first EF-hand (EF1) sequences [Citation15].
![Figure 1. Relationship between the penta-EF-hand (PEF) protein family and the calpain family in mammals.Classical (typical) calpain sequences contain the PEF domain and the calpain type β-sandwich domain (CBSW) in addition to the cysteine protease core domain (CysPc), which is further divided into two subdomains named PC1 (containing a catalytic Cys residue) and PC2 (containing catalytic His and Asn residues). Conventional calpains (μ-calpain and m-calpain) are comprised of each catalytic large subunit (designated CAPN1 for μ-calpain or CAPN2 for m-calpain) and a common regulatory small subunit (CAPNS1). Non-classical (atypical) calpain sequences lack the PEF domain but contain additional domains or motifs [calcium-binding C2 domain, microtubule-interacting and trafficking (MIT) domain, Zinc finger (ZnF), SOL-homology domain (SOH), and circularly permutated globin domain (cpGB) split by the calmodulin-binding IQ motif]. Calpain-3 (CAPN3), specifically expressed in skeletal muscles, has distinct sequences: N-terminal sequence (NS), insertion sequence 1 (IS1), and insertion sequence 2 (IS2). Calpain-7 (CAPN7) is an ortholog of fungal PalB. PEF proteins are classified into two groups based on similarity of the first EF-hand (EF1) sequences [Citation15].](/cms/asset/264e773b-f049-471a-8279-17f5315c45ff/tbbb_a_1700099_f0001_oc.jpg)
Table 1. Distribution of penta-EF-hand (PEF) proteins, classical calpains and calpain-7 orthologs in eukaryotes.
Figure 2. Schematic structures of ALG-2-interacting proteins reported in the literature.
The human or murine ALG-2-interacting proteins are classified into five groups for convenience sake based on functional properties: (a) ESCRT system, (b) ER-to-Golgi vesicular transport, (c) RNA processing, (d) protein kinases, and (e) miscellaneous. Underlined proteins have been studied in the author’s group. Red boxes and thick violet bars indicate Pro-rich regions (PRRs) and determined ALG-2-binding regions, respectively. PTP, phosphotyrosine phosphatase; UEV, ubiquitin E2 variant; CC, coiled-coil; SB, steadiness box; LC1, light chain 1; CID, C-terminal domain (CTD)-interacting domain; ZnF, zinc finger; RRM, RNA recognition motif; Ig-like C2, immunoglobulin-like constant domain type 2; ANK, ankyrin; TM, transmembrane; C2, Protein Kinase C C2-domain-like Ca2+-binding domain; vWFA, von Willebrand factor A.
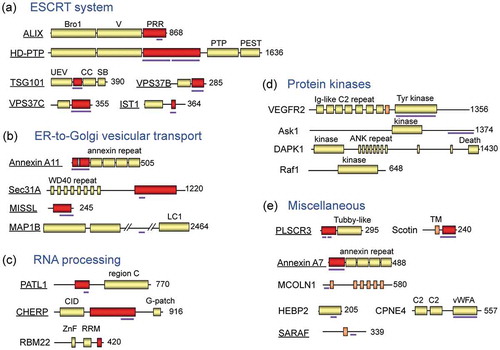
Figure 3. PEF-binding motifs and 3D structures.
(a) Three types of Pro-rich ALG-2-binding motifs (ABMs). Residues conserved among the identified ALG-2-interacting proteins in each type of ABM are indicated in red, and residues compatible with the type 2 motif at the Ω position are indicated in violet. [PΦ], Pro or hydrophobic; [FW], Phe or Trp; Ω, large side chain; x, variable. (b) Overall 3D structure of the complex between ALG-2 (homodimer) and ALIX peptides (indicated by magenta arrows) is shown by a cartoon in rainbow colors (from blue in the N-terminal region to red in the C-terminal region) using the 3D presentation software PyMOL and Protein Data Bank (PDB) code 2ZNE. (c) Overall 3D structure of the complex between ALG-2 and Sec31A peptides. PDB code 3WXA. A side view (left panel) and a 90°-rotated bottom view (right panel). (d) Schematic representation of a three-binding-site model of calpain inhibition by calpastatin. Among the three conserved regions of the four repeated domains of calpastatin, region B binds the protease domain and inhibits the proteolytic activity of calpain. Regions A and C bind the PEF domains of the large subunit (L-PEF) and the small subunit (S-PEF), respectively. (e) Amino acid sequences of regions A and C of human calpastatin. Conserved (identical or similar) residues are highlighted in light green for region A and in cyan for region C. Conserved residues between the two regions are marked with asterisks, where high conservation is indicated by bold face. (f) Overall 3D structure of the complex between rat m-calpain and calpastatin domain 1 (PDB code 3DF0). The PEF domains and the calpastatin peptide are shown by cartoon models in rainbow colors and in magenta, respectively. Other calpain domains are shown by surface representation in pale colors.
![Figure 3. PEF-binding motifs and 3D structures.(a) Three types of Pro-rich ALG-2-binding motifs (ABMs). Residues conserved among the identified ALG-2-interacting proteins in each type of ABM are indicated in red, and residues compatible with the type 2 motif at the Ω position are indicated in violet. [PΦ], Pro or hydrophobic; [FW], Phe or Trp; Ω, large side chain; x, variable. (b) Overall 3D structure of the complex between ALG-2 (homodimer) and ALIX peptides (indicated by magenta arrows) is shown by a cartoon in rainbow colors (from blue in the N-terminal region to red in the C-terminal region) using the 3D presentation software PyMOL and Protein Data Bank (PDB) code 2ZNE. (c) Overall 3D structure of the complex between ALG-2 and Sec31A peptides. PDB code 3WXA. A side view (left panel) and a 90°-rotated bottom view (right panel). (d) Schematic representation of a three-binding-site model of calpain inhibition by calpastatin. Among the three conserved regions of the four repeated domains of calpastatin, region B binds the protease domain and inhibits the proteolytic activity of calpain. Regions A and C bind the PEF domains of the large subunit (L-PEF) and the small subunit (S-PEF), respectively. (e) Amino acid sequences of regions A and C of human calpastatin. Conserved (identical or similar) residues are highlighted in light green for region A and in cyan for region C. Conserved residues between the two regions are marked with asterisks, where high conservation is indicated by bold face. (f) Overall 3D structure of the complex between rat m-calpain and calpastatin domain 1 (PDB code 3DF0). The PEF domains and the calpastatin peptide are shown by cartoon models in rainbow colors and in magenta, respectively. Other calpain domains are shown by surface representation in pale colors.](/cms/asset/0b2c982a-cb45-4253-97e1-f054755880d7/tbbb_a_1700099_f0003_oc.jpg)
Figure 4. Schematic diagram of calpain-7 actions on ESCRT-mediated EGF receptor downregulation in the endosome-to-lysosome pathway.
Calpain-7 (CAPN7) is recruited to endosomes after stimulation of cells with epidermal growth factor (EGF) and regulates downregulation of the ubiquitinated and endocytosed EGF receptor (EGFR). Calpain-7 interacts via the tandemly repeated microtubule-interacting and trafficking (MIT) domains with a subset of ESCRT-III subunits (CHMP proteins) and related proteins that contain MIT-interacting motifs (MIMs). ALG-2 interacts with IST1, a CHMP-like protein, in a Ca2+-dependent manner at the Met-Pro repeat (MP) region. Endogenous substrates of calpain-7 have not been identified yet. Fungal and yeast orthologs of calpain-7 cleave ALIX-homolog-interacting transcription factors in association with ESCRT-III proteins. VPS4 (isoforms A and B) and spastin, meiotic clade AAA ATPases containing MIT domains, disassemble ESCRT-III polymers and microtubules, respectively [Citation101,Citation102]. CHMP, charged multivesicular body protein; MTBD, microtubule-binding domain; MVB, multivesicular body; Ub, ubiquitin.
![Figure 4. Schematic diagram of calpain-7 actions on ESCRT-mediated EGF receptor downregulation in the endosome-to-lysosome pathway.Calpain-7 (CAPN7) is recruited to endosomes after stimulation of cells with epidermal growth factor (EGF) and regulates downregulation of the ubiquitinated and endocytosed EGF receptor (EGFR). Calpain-7 interacts via the tandemly repeated microtubule-interacting and trafficking (MIT) domains with a subset of ESCRT-III subunits (CHMP proteins) and related proteins that contain MIT-interacting motifs (MIMs). ALG-2 interacts with IST1, a CHMP-like protein, in a Ca2+-dependent manner at the Met-Pro repeat (MP) region. Endogenous substrates of calpain-7 have not been identified yet. Fungal and yeast orthologs of calpain-7 cleave ALIX-homolog-interacting transcription factors in association with ESCRT-III proteins. VPS4 (isoforms A and B) and spastin, meiotic clade AAA ATPases containing MIT domains, disassemble ESCRT-III polymers and microtubules, respectively [Citation101,Citation102]. CHMP, charged multivesicular body protein; MTBD, microtubule-binding domain; MVB, multivesicular body; Ub, ubiquitin.](/cms/asset/c935265e-de3b-434f-bc1c-af6163fbabfa/tbbb_a_1700099_f0004_oc.jpg)
