Figures & data
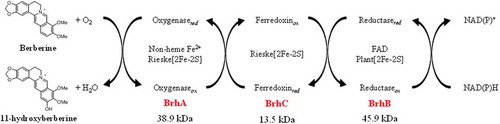
Figure 1. Berberine and palmatine metabolism by berberine-utilizing bacteria. BBR is degraded into D-BBR as the common BBR metabolite in four distinct strains. As a gene that cleaves the methylenedioxy ring of BBR, BD7100 and GBD-1 have a brdA, and BD3100 and CJ1 have two homologous genes, brdA1 and brdA2. BD3100 and CJ1 generated a BBR 11-hydroxylation product, and BD3100 has a PAL 11-hydroxylation pathway. The 11-hydroxylation pathway of BBR is the focus of this study. BBR, berberine; D-BBR, demethyleneberberine; H-BBR, 11-hydroxyberberine; HD-BBR, 11-hydroxydemethyleneberberine; HDBA, 2-hydroxy-3,4-dimethoxybenzeneacetic acid; PAL, palmatine; H-PAL, 11-hydroxypalmatine. Solid arrows indicate the degradation pathway by BBR-utilizing bacteria, and the dashed line indicates the deduced degradation pathway. Arrows with multiple heads indicates multiple metabolic stages.
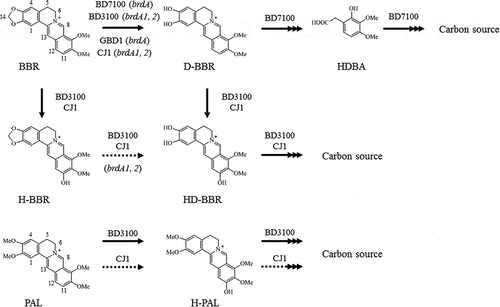
Figure 2. The growth of, and berberine degradation by, strain CJ1 and its mutants. (a) Growth of CJ1 (Wile-type; WT) and the mutants Δ7331, Δ7332 and Δ7340 after 5 days on 0.1 × LB (Luria-bertani) medium containing 1 mM BBR. (b, c) The growth (as assessed by optical density) of CJ1 and mutants in (b) LB medium and (c) minimal medium containing 1 mM berberine (BBR). Symbols: CJ1 (white circles), Δ7331 (black squares), Δ7332 (black triangles), and Δ7340 (black circles).
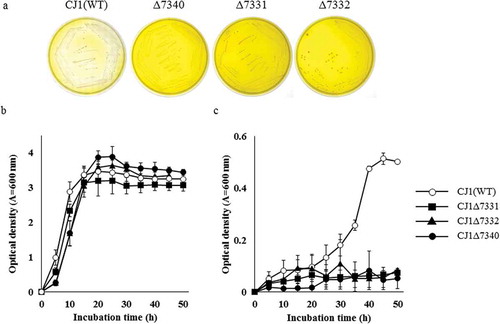
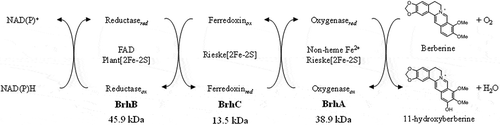
Figure 3. Alignment of the predicted amino acid sequence of BrhA, BrhB, and BrhC with the sequences of paralogs. Alignment of BrhA with DdmC and CndA shows the conserved domain (shadow box) in this class. Proteins in this family contain a Rieske domain with the consensus sequence Cys-X-His (CXH) and Cys-X-X-His (CX2H) and a non-heme iron domain with the consensus sequence Asp-X-X-X-Asp-X-X-His-X-X-X-X-His (DX3DX2HX4H). BrhA (LC500707), oxygenase component of BBR 11-hydroxylase from Burkholderia sp. CJ1 (BurkCJ1); CndA (KJ461679), oxygenase component of N-dealkylation of chloroacetanilide herbicides from Sphingomonads DC-6 (SphiDC6); DdmC (AY786443), oxygenase component of dicamba O-demethylase from Stenotrophomonas maltophilia strain DI-6 (SmalDI6). (b) Alignment of BrhB with Glutathione type (GR-type) reductases shows the conserved domain (shadow box) in this class. Proteins in this family have two ADP-binding domains (for FAD and NADH, respectively) with the consensus sequence Gly-X-Gly-X-X-Gly-X-X-X-Ala (GXGX2GX3A) and a flavin-binding domain (for FAD) with the consensus sequence Thr-X-X-X-X-X-X-Ala-X-Gly-Asp (TX6AXGD). BrhB (LC500708), reductase component of BBR 11-hydroxylase from BurkCJ1; DdmA1 (AY786444) and DdmA2 (AY786445), reductase component of DdmC from SmalDI-6; CndC1 (AHW42445), reductase component of CndA from SphiDC6. (c) Alignment of BrhC with plant-type ferredoxins shows the conserved domain (shadow box) in this class. Proteins in this family contain a [2Fe-2S] domain with the consensus sequence Cys-X-X-X-X-X-Cys-X-X-Cys-(inserted 36 or 37 of any amino acid; X)-Cys (CX5CX2CX36/37C). BrhC, ferredoxin component of BrhC (LC500709) from BurkCJ1; CndB1 (AHW42448) and CndB2 (AHW42449); ferredoxin components of from SphiDC-6; DdmB (AAV53698), ferredoxin component of DdmC from SmalDI-6 (GenBank accession numbers in parentheses).
![Figure 3. Alignment of the predicted amino acid sequence of BrhA, BrhB, and BrhC with the sequences of paralogs. Alignment of BrhA with DdmC and CndA shows the conserved domain (shadow box) in this class. Proteins in this family contain a Rieske domain with the consensus sequence Cys-X-His (CXH) and Cys-X-X-His (CX2H) and a non-heme iron domain with the consensus sequence Asp-X-X-X-Asp-X-X-His-X-X-X-X-His (DX3DX2HX4H). BrhA (LC500707), oxygenase component of BBR 11-hydroxylase from Burkholderia sp. CJ1 (BurkCJ1); CndA (KJ461679), oxygenase component of N-dealkylation of chloroacetanilide herbicides from Sphingomonads DC-6 (SphiDC6); DdmC (AY786443), oxygenase component of dicamba O-demethylase from Stenotrophomonas maltophilia strain DI-6 (SmalDI6). (b) Alignment of BrhB with Glutathione type (GR-type) reductases shows the conserved domain (shadow box) in this class. Proteins in this family have two ADP-binding domains (for FAD and NADH, respectively) with the consensus sequence Gly-X-Gly-X-X-Gly-X-X-X-Ala (GXGX2GX3A) and a flavin-binding domain (for FAD) with the consensus sequence Thr-X-X-X-X-X-X-Ala-X-Gly-Asp (TX6AXGD). BrhB (LC500708), reductase component of BBR 11-hydroxylase from BurkCJ1; DdmA1 (AY786444) and DdmA2 (AY786445), reductase component of DdmC from SmalDI-6; CndC1 (AHW42445), reductase component of CndA from SphiDC6. (c) Alignment of BrhC with plant-type ferredoxins shows the conserved domain (shadow box) in this class. Proteins in this family contain a [2Fe-2S] domain with the consensus sequence Cys-X-X-X-X-X-Cys-X-X-Cys-(inserted 36 or 37 of any amino acid; X)-Cys (CX5CX2CX36/37C). BrhC, ferredoxin component of BrhC (LC500709) from BurkCJ1; CndB1 (AHW42448) and CndB2 (AHW42449); ferredoxin components of from SphiDC-6; DdmB (AAV53698), ferredoxin component of DdmC from SmalDI-6 (GenBank accession numbers in parentheses).](/cms/asset/46cb831c-fe56-41ca-b444-8dd82b469c38/tbbb_a_1722056_f0003_b.gif)
Figure 4. Investigation of brhA, brhB, and brhC involvement in berberine 11-hydroxylation using a resting-cell assay. The resting-cell assay of CJ1 (wild type; WT) and the mutants CJ1Δ7331, CJ1Δ7332, and CJ1Δ7340. The curves indicate the results obtained with (a) CJ1, (b) CJ1Δ7331, (c) CJ1Δ7332, and (d) CJ1Δ7340. Each curve shows the remaining concentration of BBR (100% with 0.5 mM berberine added) indicated by black circles and the peak areas of D-BBR (black squares), H-BBR (white circles), and HD-BBR (white squares) during the resting-cell assay. BBR, berberine; D-BBR, demethyleneberberine; H-BBR, 11-hydroxyberberine; HD-BBR, 11-hydroxydemethyleneberberine. (e) HPLC chromatograms (280 nm) of resting-cell assay with BBR. CJ1(WT) degrades BBR to H-BBR and HD-BBR after 3 h in the assay.
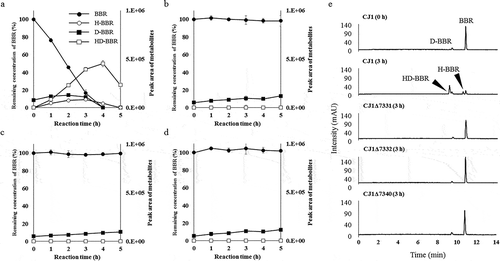
Figure 5. Examination of BBR 11-hydroxylation ability of brhA by complementation experiment. (a) Growth of control strain (CJ1Δ7340/pJB861) and complemented strain (CJ1Δ7340/pJB7340) after 5 days on 0.1 × LB medium containing 1 mM berberine (BBR). (b) Each curves shows the remaining concentration of BBR (100% with 0.5 mM BBR added) and the peak areas of D-BBR, H-BBR, and HD-BBR. Curves indicating the results obtained with the control (BBR: black circles, D-BBR: black squares, H-BBR: black diamonds, HD-BBR: black triangles) and complemented strains (BBR: white circles, D-BBR: white squares, H-BBR: white diamonds, HD-BBR: white triangles) during the resting-cell assays. D-BBR, demethyleneberberine; H-BBR, 11-hydroxyberberine; HD-BBR, 11-hydroxydemethyleneberberine. (c) HPLC chromatograms (280 nm) of resting-cell assay with BBR. Complemented strain degraded BBR to H-BBR and HD-BBR after 1 h in the assay.
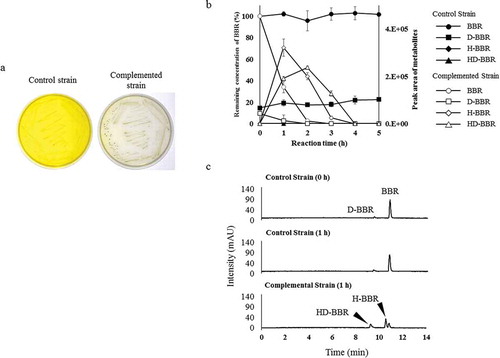
Figure 6. Purified-enzyme assays under various conditions. (a) Curves indicating the results of a purified-enzyme assay including all 3 enzymes in the presence of NADH and Fe2+. Each curve shows the remaining concentration of BBR (100% with 0.5 mM BBR added) and the peak area of H-BBR. Curves indicating the control (BBR: black circles, H-BBR: black squares) and enzyme added (BBR: white circles, H-BBR: white squares). BBR, berberine; H-BBR, 11-hydroxyberberine. (b) HPLC chromatograms (280 nm) of the reactions at 60 min in the presence of various combinations of purified enzymes; all reactions were performed in the presence of 0.5 mM NADH and 1 mM Fe2+. The BBR standard is shown at the bottom. The production of H-BBR was observed only with the combination of BrhA, BrhB, and BrhC in the presence of NADH and Fe2+.
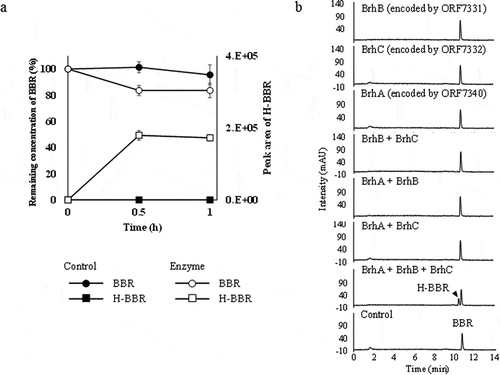
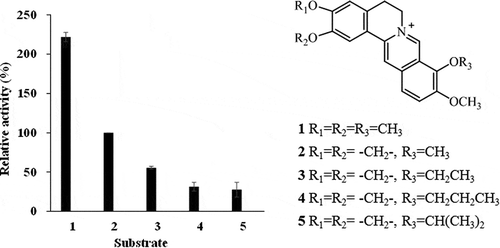
Figure 7. Examination of substrate specificity of recombinant berberine 11-hydroxylase protein. The bar numbers in the graph indicate the compound numbers of the tested substrates: 1, palmatine (PAL); 2, berberine (BBR); 3, 9-O-ethyl berberine (9-O-ethyl BBR); 4, 9-O-propyl berberine (9-O-propyl BBR); and 5, 9-O-isopropyl berberine (9-O-isopropyl BBR). The relative activity was calculated by normalizing 11-hydroxyberberine (H-BBR) production at 60 min against that obtained with BBR (2).