Figures & data
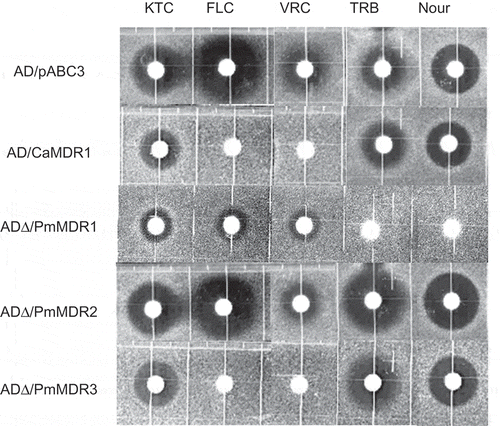
Table 1. Yeast strains used in this study.
Figure 1. Cloning strategy for PmMDR1, PmMDR2 and PmMDR3 in Saccharomyces cerevisiae ADΔ using the yeast membrane protein hyperexpression system.The transformation cassette consists of a 1.1-kb DNA fragment (upstream region of PDR5), open reading frame (ORF) to be cloned, and a 1.7-kb DNA fragment (PGK1 terminator, URA3 marker and downstream end of the PDR5 ORF (ds)). The PmMDR1, PmMDR2, and PmMDR3 ORFs were PCR amplified using primer sets as shown in the figure and Supplementary Table S1, employing either cDNA (PmMDR1 and PmMDR2) or genomic DNA (PmMDR3) as templates. For cloning of PmMDR2 and PmMDR3 the amplified ORF was inserted in plasmid pABC3 between the PacI and NotI sites and then amplified in E. coli DH5α; the transformation cassette then was excised from the newly constructed plasmid by AscI digestion. For cloning of PmMDR1, the ORF was directly used for transformation with the 1.1-kb and 1.7-kb DNA fragments, which were PCR amplified using pABC3 as template and primer pairs of AscI_for and primer C (PDR5up_RP.034610) and AscI_rev and primer D (PGK1_FP_034610) (Lamping et al., 2007). For each of the 3 PmMDR genes, the resulting transformation cassette was integrated into the S. cerevisiae AD∆ chromosome at the PDR5 locus by homologous recombination. Primers used for the individual genes were as follows (A and B, respectively): for PmMDR1, PMAA_034610_F1 and PMAA_034610_R; for PmMDR2, PMAA_080940-FP and PMAA_080940-RP; for PmMDR3, Fwd-067100-MFS 0984413–1 and Rev-067100-MFS 0984287–1.
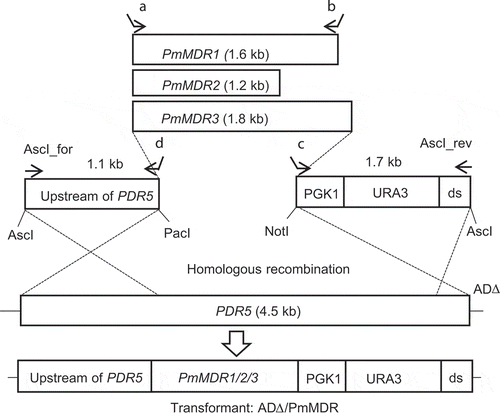
Table 2. Classification of typical P. marneffei MFS transporters with 12 and 14 transmembrane spans (TMSs).
Figure 2. The phylogenetic relationship of all characterized DHA1 transporters in S. cerevisiae, C. albicans, C. glabrata, C. neoformans, and P. marneffei.The proteins characterized in the present work (PmMDR1, PmMDR2, and PmMDR3) are indicated in boldfaced type.
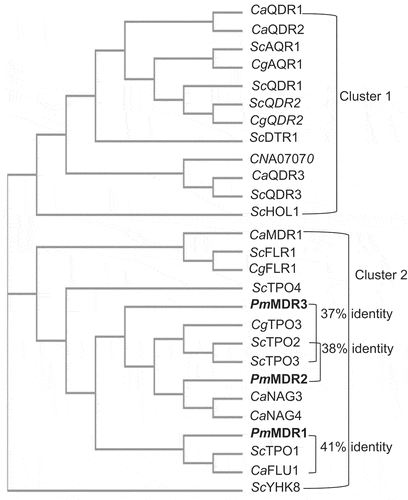
Figure 3. Predicted topology of fungal DHA1-subfamily transporters, including PmMDR1, PmMDR2, and PmMDR3, showing major topological features and motifs.Glutamate (e) and aspartate (d) in black boxes are important residues for interactions with polyamines [Citation22].
![Figure 3. Predicted topology of fungal DHA1-subfamily transporters, including PmMDR1, PmMDR2, and PmMDR3, showing major topological features and motifs.Glutamate (e) and aspartate (d) in black boxes are important residues for interactions with polyamines [Citation22].](/cms/asset/e1912e83-b727-4906-a3e8-28b3e83267eb/tbbb_a_1732185_f0003_b.gif)
Table 3. Drug susceptibilities of S. cerevisiae AD∆ overexpressing P. marneffei PmMDR1, PmMDR2, and PmMDR3, and C. albicans CaMDR1.
Figure 5. Susceptibility of S. cerevisiae ADΔ/pABC3, ADΔ/CaMDR1, AD∆/PmMDR1, AD∆/PmMDR2, and AD∆/PmMDR3 to xenobiotics.Amounts (μg) loaded on discs: KTC, ketoconazole (0.04); FLC, fluconazole (3); VRC, voriconazole (0.025); TRB, terbinafine (100); Nour, nourseothricin (50).
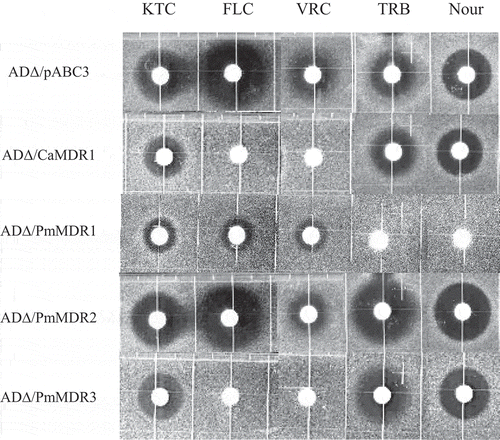
Table.supplement.S1.pdf
Download PDF (266.1 KB)Data availability statement
The data described in this article are openly available in the Open Science Framework at DOI:10.17605/OSF.IO/TPA6U.