Figures & data
Table 1. The correlation between ACTL8 expression and the clinic-pathological factors of patients with EC from TCGA database.
Figure 1. ACTL8 expression was enhanced in EC and predictive of worse prognosis. (a) The expression level of ACTL8 in 35 normal human endometrium samples and 552 EC samples was analyzed based on the RNA-seq data downloaded from TCGA database. (b) Kaplan–Meier analysis was performed to determine the correlation between ACTL8 expression and overall survival of patients with EC. The 541 samples with complete clinical data were grouped into high (n = 270) and low (n = 271) expression groups according to the median of ACTL8 expression level. (c). The mRNA expression of ACTL8 in HEC-1-A, KLE, Ishikawa, and NUEEC cells was detected by qRT-PCR. n = 6; *p < 0.05, **p < 0.01 vs. NUEEC group.
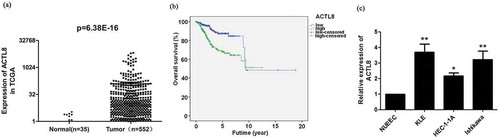
Table 2. Cox proportional hazards model analysis of clinic pathologic features for overall survival in patients with EC from TCGA database.
Figure 2. ACTL8 expression in EC cell lines and knockdown efficiency detection. (a–b) The mRNA and protein expression of ACTL8 in KLE cells transfected with PLKO.1 (control) or PLKO.1-sh ACTL8 (sh ACTL8) was detected by qRT-PCR (a) and western blot (b). (c–d) The mRNA and protein expression of ACTL8 in Ishikawa cells transfected with PLKO.1 (control) or PLKO.1-sh ACTL8 (sh ACTL8) was detected by qRT-PCR (c) and western blot (d). n = 6; **p < 0.01 vs. control group, ##p < 0.01 vs. sh ACTL8-1 group.
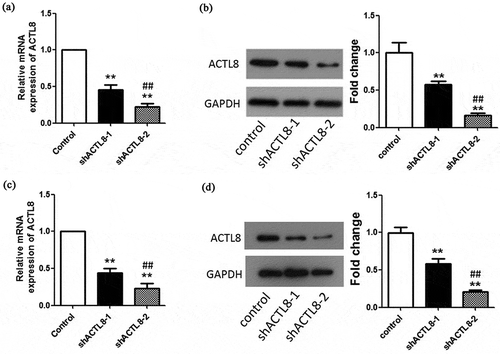
Figure 3. Knockdown of ACTL8 repressed EC cell proliferation. (a, d) The proliferative curves of KLE (a) and Ishikawa (d) cells transfected with PLKO.1 (control) or PLKO.1-sh ACTL8 (sh ACTL8). (b, e) The colony formation ability of KLE (b) and Ishikawa (e) cells was determined using a colony formation assay. (c, f) The number of KLE (c) and Ishikawa (f) clones formed in control and sh ACTL8 groups. n = 6; **p < 0.01 vs. control group.
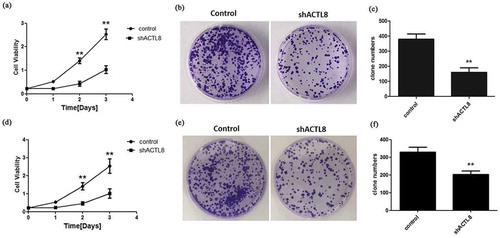
Figure 4. Knockdown of ACTL8 repressed EC cell invasion and migration. (a, c) Representative images of the invaded and migrated KLE (a) and Ishikawa (c) cells were captured by using an inverted microscope at magnification×200. (b, d) The numbers of invaded and migrated KLE (b) and Ishikawa (d) cells were detected by transwell assays. n = 6; **p < 0.01 vs. control group.
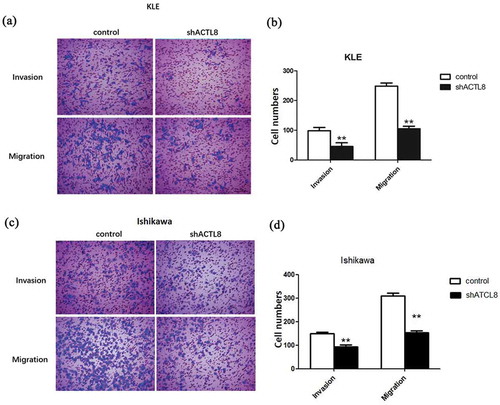
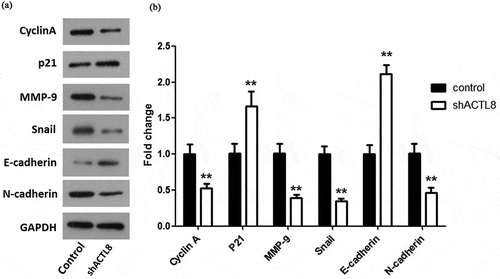
Figure 5. Knockdown of ACTL8 up-regulated the expression of p21, E-cadherin, and down-regulated the expression of Cyclin A, MMP-9, Snail, and N-cadherin. (a) The expression of Cyclin A, p21, MMP-9, Snail, E-cadherin, and N-cadherin in KLE cells transfected with PLKO.1 (control) or PLKO.1-sh ACTL8 (sh ACTL8) were detected by western blot. (b) Quantification of the protein expression levels in Figure (a). n = 6; **p < 0.01 vs. control group.